Integration use case: post-analysis of sequences with immunarch¶
This use case will show how to perform a post-analysis of CMV associated TCRβ sequences using immunarch/ These sequences were identified through the method published by Emerson et al., which was reproduced inside immuneML. The analysis used to obtain these sequences is described in Manuscript use case 1: Reproduction of a published study inside immuneML, where the sequences were exported using the RelevantSequenceExporter.
Download the sequence file here: relevant_sequences.csv
This analysis requires the R package immunarch version 0.6.5 or higher to be installed.
library(immunarch)
# Read in of CMV-associated sequences
relevant_sequences <- read.csv("relevant_sequences.csv") # Column names are AIRR-compliant
relevant_sequences = relevant_sequences[!(relevant_sequences$v_call==""), ] # Remove sequences with missing V gene annotation
colnames(relevant_sequences) <- c("CDR3.aa", "V.name", "J.name") # Renaming of columns in order for geneUsage function to work properly
relevant_sequences$Clones <- 1 # Addition of clones columns in order for geneUsage function to work properly
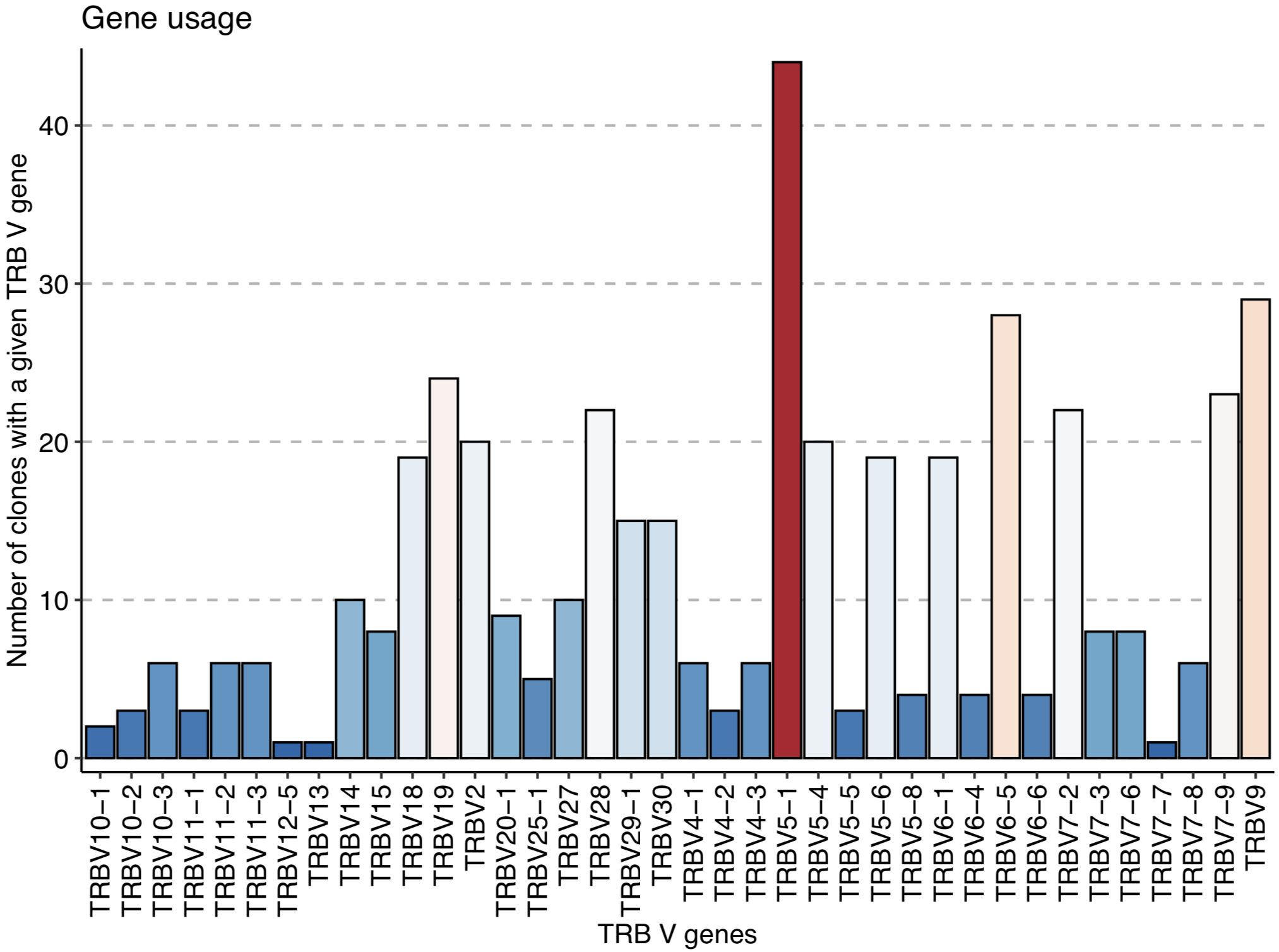
# V gene analysis using immunarch:
geneUsage(relevant_sequences) %>% vis() + labs (x = "TRB V genes", y = "Number of clones with a given TRB V gene")
The immunarch geneUsage() plot shows that TRBV5-1 is the most used V gene among the CMV-associated TCRβ sequences: