Welcome to the former immuneML documentation!¶
immuneML is a platform for machine learning-based analysis and classification of adaptive immune receptors and repertoires (AIRR). To get started using immuneML right away, check out our Quickstart tutorial.
immuneML can be used for:
Training ML models for repertoire classification (e.g., disease prediction) or receptor sequence classification (e.g., antigen binding prediction). In immuneML, the performance of different machine learning (ML) settings can be compared by nested cross-validation. These ML settings consist of data preprocessing steps, encodings and ML models and their hyperparameters.
Exploratory analysis of datasets by applying preprocessing and encoding, and plotting descriptive statistics without training ML models.
Simulating immune events, such as disease states, into experimental or synthetic repertoire datasets. By implanting known immune signals into a given dataset, a ground truth benchmarking dataset is created. Such a dataset can be used to test the performance of ML settings under known conditions.
Applying trained ML models to new datasets with unknown class labels.
The starting point for any immuneML analysis is the YAML specification file. In this file, the settings of the analysis components are defined (also known as definitions), which are shown in six different colors in the figure below. Additionally, the YAML file describes one or more instructions, which corresponds to one of the applications listed above (and some additional instructions).
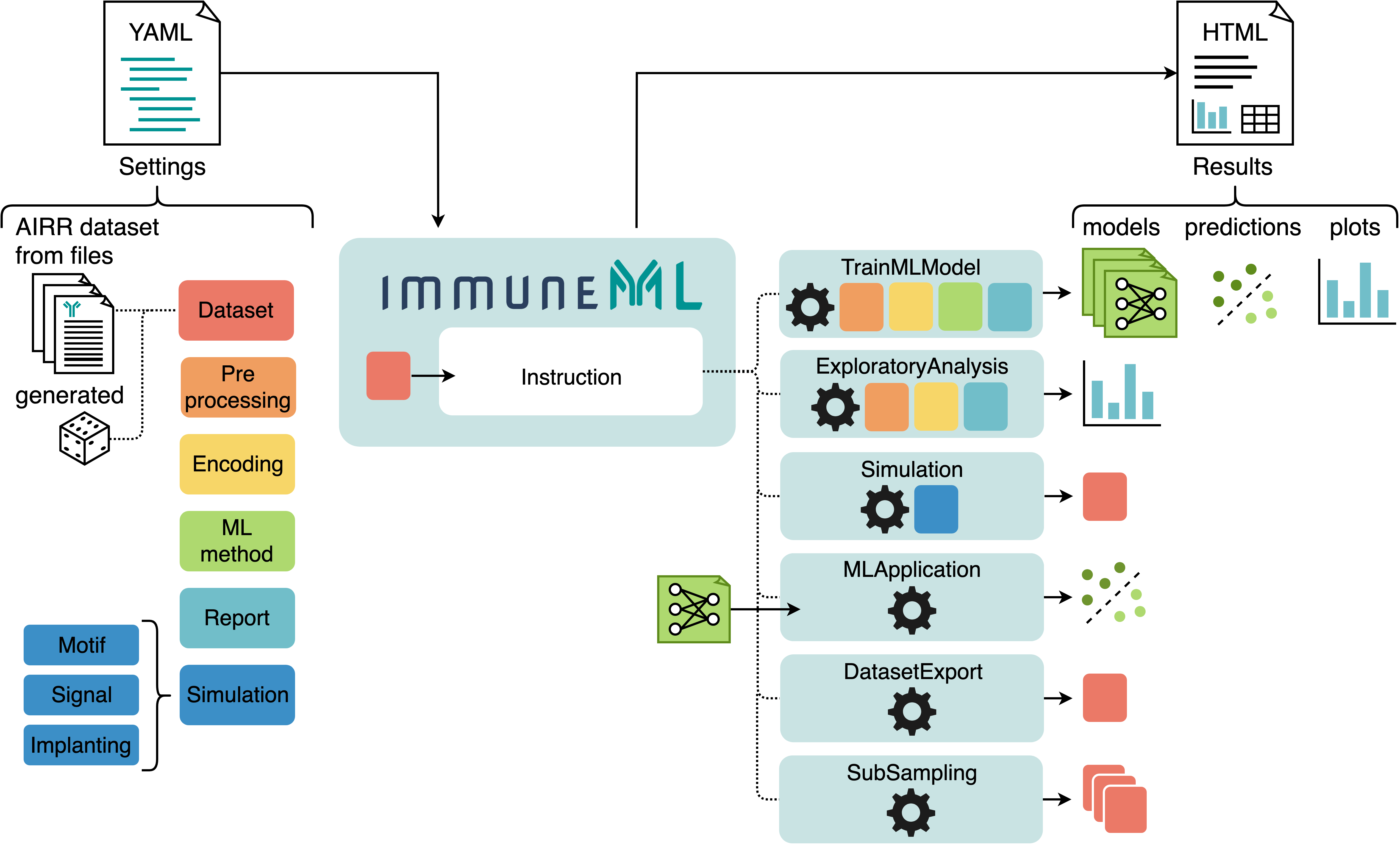
An overview of immuneML usage: analysis components and instructions are specified in a YAML file. Each use case corresponds to a different instruction. The results of the instructions are summarized and presented in an HTML file.¶
If you want to use immuneML locally, see Installing immuneML.
Alternatively, to run immuneML in a web browser, go to our Galaxy Portal. Here, we offer the same functionalities as in the command-line interface, and in addition simplified button-based interfaces for training classifiers.
To become familiar with the YAML-based specification, you can find a concrete example in our Quickstart guide, or read about the overall YAML structure and options in How to specify an analysis with YAML.
YAML specifications are equivalent between the Galaxy and command-line interfaces. However, in Galaxy datasets must first be converted to a dataset collection (see How to make an immuneML dataset in Galaxy).
We offer Tutorials for specific use cases (e.g., how to train and assess classifiers, or how to generate synthetic immune repertoire data for benchmarking purposes), and under immuneML & Galaxy you will find tutorials using Galaxy as a starting point.
If you are wondering about all the possible analysis components and their options, you can find the complete list and documentation under YAML specification.
Our open-source code can be found on GitHub :)