How to train and assess a receptor or repertoire-level ML classifier¶
immuneML provides a rich set of functionality for training and assessing machine learning models to classify of receptors or repertoires. This is done using the TrainMLModel instruction. The goal of the training process is to learn the parameters for a given ML model that result in the most accurate predictions. Furthermore, we want to select the optimal settings for hyperparameters, which are tunable characteristics of an ML model which influence the training process and can not automatically be learned by training.
Aside from the hyperparameters of the ML models themselves, the choices made for preprocessing or filtering of the dataset, as well as the encoding and its parameters will influence the performance of the trained ML model. Therefore, preprocessing steps and encoding are treated as hyperparameters during ML model training, enabling automatic optimization and unbiased assessment of these choices.
To learn the parameters and hyperparameters of the ML model, the data needs to be split into training, validation and test sets. Models with different hyperparameters are trained on the training set, then assessed on the separate validation set to find the optimal hyperparameters, and finally tested on the test set to estimate the model performance. The resulting optimized classifier can also afterwards be applied to further datasets (see: How to apply previously trained ML models to a new dataset). This process is shown in the figure below:
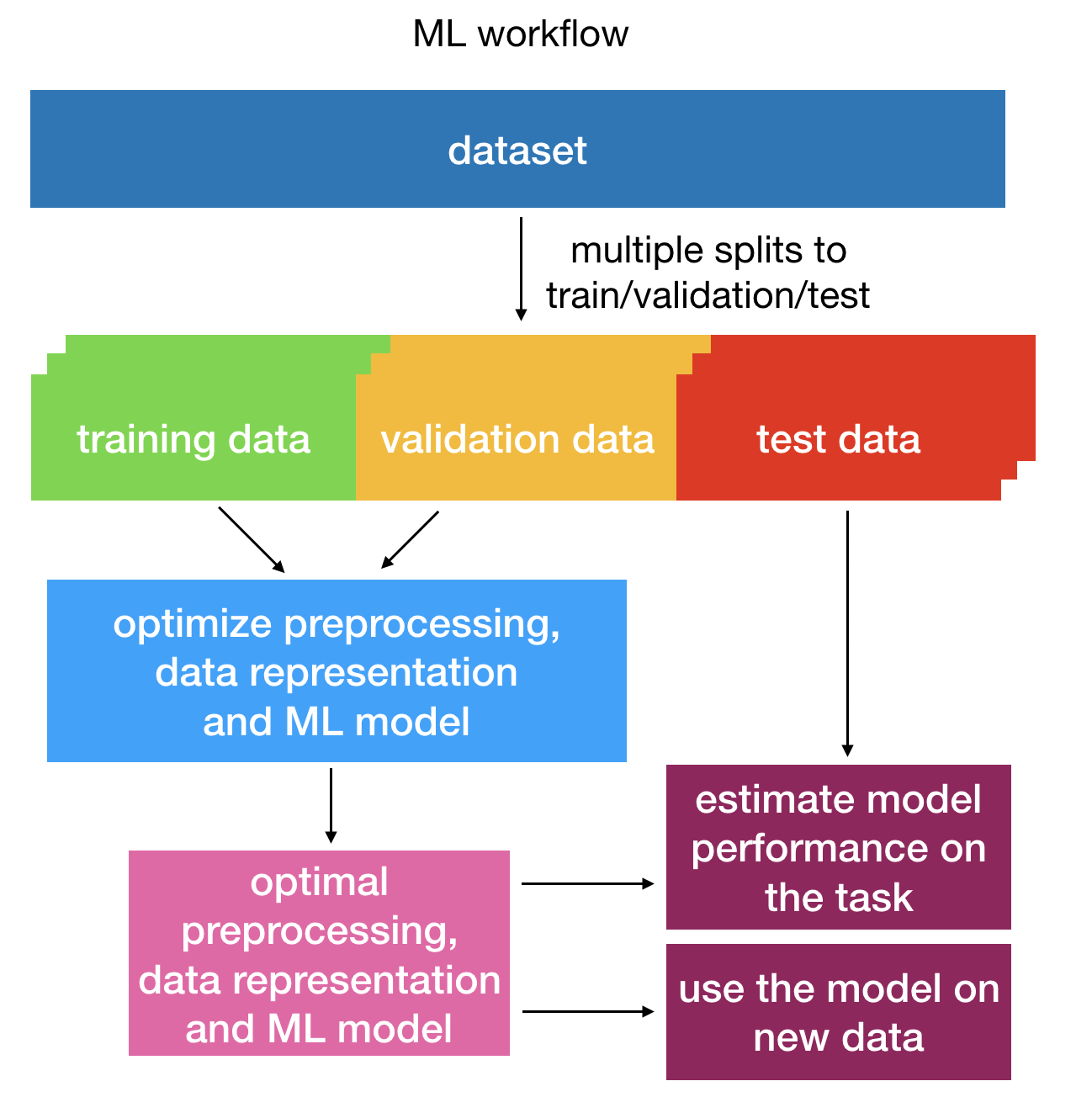
Overview of the training process of an ML classifier: hyperparameter optimization is done on training and validation data and the model performance is assessed on test data¶
See also How to discover disease- or antigen specificity-associated motifs to read more about ML interpretability.
YAML specification of TrainMLModel¶
Definitions section¶
When running the TrainMLModel instruction, the definitions part of the YAML specification should at
least contain datasets, encodings and ml_methods components. Optionally, preprocessing_sequences
and reports may be defined. For detailed descriptions of how the parameters for each of these components are defined,
please refer to the YAML specification documentation. Under Definitions each analysis component is documented
and settings are described.
For importing a dataset, see How to import data into immuneML and for more details on writing the YAML specification, see How to specify an analysis with YAML.
An example of the definitions section using a repertoire dataset is given here:
definitions:
datasets: # every instruction uses a dataset
my_dataset:
format: AIRR
params:
path: path/to/repertoires/
metadata_file: path/to/metadata.csv
preprocessing_sequences:
my_preprocessing: # user-defined preprocessing name
- my_beta_chain_filter:
ChainRepertoireFilter:
keep_chain: TRB
ml_methods:
my_svm: # example ML method with user-defined settings
SVM: # Here, a third level of 3-fold cross-validation is used
penalty: # to determine the optimal hyperparameters for 'C' and 'penalty'
- l1 # This functionality is only available for scikit-learn classifiers
- l2
C:
- 0.01
- 0.1
- 1
- 10
- 100
model_selection_cv: True
model_selection_n_folds: 3
my_log_reg: LogisticRegression # example ML method with default settings
encodings:
my_kmer_freq_encoding:
KmerFrequency:
k: 3
reports:
my_coefficients: Coefficients
my_sequence_lengths: SequenceLengthDistribution
my_performance: MLSettingsPerformance
When choosing which ML method(s) are most suitable for your use-case, please consider the following table. The table describes which of the ML methods can be used for binary classification (two classes per label), and which can be used for multi-class classification. Note that all classifiers can automatically be used for multi-label classification in immuneML. Furthermore, it describes what type of dataset the classifier can be applied to, and whether a third level of nested cross-validation can be used for the selection of model parameters (scikit-learn classifiers).
ML method |
binary classification |
multi-class classification |
sequence dataset |
receptor dataset |
repertoire dataset |
model selection CV |
|---|---|---|---|---|---|---|
AtchleyKmerMILClassifier |
✓ |
✗ |
✗ |
✗ |
✓ |
✗ |
DeepRC |
✓ |
✗ |
✗ |
✗ |
✓ |
✗ |
KNN |
✓ |
✓ |
✓ |
✓ |
✓ |
✓ |
LogisticRegression |
✓ |
✓ |
✓ |
✓ |
✓ |
✓ |
ProbabalisticBinaryClassifier |
✓ |
✗ |
✗ |
✗ |
✓ |
✗ |
RandomForestClassifier |
✓ |
✓ |
✓ |
✓ |
✓ |
✓ |
ReceptorCNN |
✓ |
✗ |
✗ |
✓ |
✗ |
✗ |
SVM |
✓ |
✓ |
✓ |
✓ |
✓ |
✓ |
TCRdistClassifier |
✓ |
✓ |
✓ |
✓ |
✓ |
✗ |
Instructions section¶
The YAML specification of the TrainMLModel instruction contains various different components.
Firstly, we should give our instruction a unique name and set the type:
instructions:
my_instr:
type: TrainMLModel
... # other parameters should be added here
Furthermore, we should define which dataset to train the models on, and which labels to predict.
The labels are defined based on the metadata file (repertoire datasets) or metadata column mapping (sequence or receptor datasets),
see How to import data into immuneML for details.
In most cases there will only be one label, but it is possible to specify multiple labels in order to train
multiple different classifiers.
dataset: my_dataset
labels:
- disease_1
- disease_2
In the settings section the different combinations of ML settings must be specified. They consist of
an encoding, an ml_method and optional preprocessing (only available for repertoire datasets),
which should be referenced by the names that were used to defined them under definitions.
Not every combination of encodings and ML methods is valid. For all the valid options, see the figure in the YAML specification documentation. An example of the settings section of the instruction is:
settings:
- encoding: my_kmer_freq_encoding # Testing my_kmer_freq_encoding in combination with
ml_method: my_log_reg # my_log_reg and my_svm, with and without my_preprocessing
- encoding: my_kmer_freq_encoding
ml_method: my_svm
- preprocessing: my_preprocessing # preprocessing can only be defined for repertoire datasets
encoding: my_kmer_freq_encoding
ml_method: my_log_reg
- preprocessing: my_preprocessing
encoding: my_kmer_freq_encoding
ml_method: my_svm
Nested cross-validation consists of two loops:
the inner
selectionloop, which is used to select the optimal hyperparameter settings using the validation set(s)the outer
assessmentloop, which is used to give an unbiased assessment of the trained models using the test set(s)
Both for the selection and assessment loops, we need to define what splitting strategy should be used for the data.
The parameters that should be defined under selection and assessment are described in the SplitConfig
documentation.
Optionally, it is possible to specify various types of reports here (for more details see ReportConfig):
models: ML model reports which will be applied to all compatible models in the given loop.encoding: Encoding reports which will be applied to the encoded data of a given split (all data or training+validation data)data: Data reports which will be applied to the complete dataset of a given split (all data or training+validation data)data_splits: Data reports which will be applied to the data splits (training/validation or training+validation/test splits)
The following example shows a piece of the YAML specification when two different splitting strategies are used for both loops:
assessment: # example defining 5-fold cross-validation
split_strategy: k_fold
split_count: 5
reports:
models: # plot the coefficients of the trained models
- my_coefficients
data: # run this report on all data
- my_sequence_lengths
selection: # example defining 3-fold monte carlo cross-validation with
split_strategy: random # 70% randomly chosen training data per split
split_count: 3
training_percentage: 0.7
reports: # run this report on training/validation splits
data_splits:
- my_sequence_lengths
Furthermore, the optimization_metric and search strategy (used to determine optimal hyperparameters) must be set.
Currently, GridSearch is the only supported search strategy. Aside from the optimization_metric, other performance
metrics can also be computed for reference. And the user needs to decide whether the optimal ML settings should be
retrained on the complete dataset (including test set) after assessment through setting the parameter refit_optimal_model.
The following YAML example shows how these parameters are defined:
optimization_metric: balanced_accuracy # the metric used for optimization
metrics: # other metrics to compute
- accuracy
- auc
strategy: GridSearch
refit_optimal_model: False
Train ML model reports summarizing the performance of multiple different models should be set under reports.
And finally, the number_of_processes used for parallelization should be set.
reports:
- my_performance
number_of_processes: 4
Complete YAML¶
An example of the complete YAML specification is shown here:
definitions:
datasets: # every instruction uses a dataset
my_dataset:
format: AIRR
params:
path: path/to/repertoires/
metadata_file: path/to/metadata.csv
preprocessing_sequences:
my_preprocessing: # user-defined preprocessing name
- my_beta_chain_filter:
ChainRepertoireFilter:
keep_chain: TRB
ml_methods:
my_svm: # example ML method with user-defined settings
SVM: # Here, a third level of 3-fold cross-validation is used
penalty: # to determine the optimal hyperparameters for 'C' and 'penalty'
- l1 # This functionality is only available for scikit-learn classifiers
- l2
C:
- 0.01
- 0.1
- 1
- 10
- 100
model_selection_cv: True
model_selection_n_folds: 3
my_log_reg: LogisticRegression # example ML method with default settings
encodings:
my_kmer_freq_encoding:
KmerFrequency:
k: 3
reports:
my_coefficients: Coefficients
my_sequence_lengths: SequenceLengthDistribution
my_performance: MLSettingsPerformance
instructions:
my_instr:
type: TrainMLModel
dataset: my_dataset
labels:
- disease_1
- disease_2
settings:
- encoding: my_kmer_freq_encoding # Testing my_kmer_freq_encoding in combination with
ml_method: my_log_reg # my_log_reg and my_svm, with and without my_preprocessing
- encoding: my_kmer_freq_encoding
ml_method: my_svm
- preprocessing: my_preprocessing # preprocessing can only be defined for repertoire datasets
encoding: my_kmer_freq_encoding
ml_method: my_log_reg
- preprocessing: my_preprocessing
encoding: my_kmer_freq_encoding
ml_method: my_svm
assessment: # example defining 5-fold cross-validation
split_strategy: k_fold
split_count: 5
reports:
models: # plot the coefficients of the trained models
- my_coefficients
data: # run this report on all data
- my_sequence_lengths
selection: # example defining 3-fold monte carlo cross-validation with
split_strategy: random # 70% randomly chosen training data per split
split_count: 3
training_percentage: 0.7
reports: # run this report on training/validation splits
data_splits:
- my_sequence_lengths
optimization_metric: balanced_accuracy # the metric used for optimization
metrics: # other metrics to compute
- accuracy
- auc
strategy: GridSearch
refit_optimal_model: False
reports:
- my_performance
number_of_processes: 4
Example datasets¶
Below you will find example datasets that can be used to test out the TrainMLModel instruction.
Repertoire dataset¶
An example dataset for testing out repertoire classification in immuneML is the Quickstart dataset: quickstart_data.zip
This is a dataset in AIRR format and can be imported as follows:
definitions:
datasets: # every instruction uses a dataset
my_dataset:
format: AIRR
params:
path: path/to/repertoires/
metadata_file: path/to/metadata.csv
For this dataset, the label that can be used for prediction is ‘signal_disease’.
Sequence dataset¶
An example dataset for sequence classification of epitope GILGFVFTL can be downloaded here: sequences.tsv.
To import this dataset, use the following YAML snippet:
definitions:
datasets: # every instruction uses a dataset
my_dataset:
format: AIRR
params:
path: path/to/sequences.tsv
is_repertoire: false
paired: false
metadata_column_mapping:
epitope: epitope
For this dataset, the label that can be used for prediction is ‘epitope’.
Receptor dataset¶
An example dataset for receptor classification of epitope GILGFVFTL can be downloaded here: receptors.tsv
To import this dataset, use the following YAML snippet:
definitions:
datasets: # every instruction uses a dataset
my_dataset:
format: AIRR
params:
path: path/to/receptors.tsv
is_repertoire: false
paired: true
receptor_chains: TRA_TRB
metadata_column_mapping:
epitope: epitope
For this dataset, the label that can be used for prediction is ‘epitope’.