Manuscript use case 3: Benchmarking ML methods on ground-truth synthetic data
In this use case, we show that immuneML can be used for benchmarking ML methods and encodings. To do this, we first simulate ground-truth synthetic adaptive immune repertoires, then implant known disease signals in these repertoires of varying difficulties, and finally perform a benchmarking.
The complete collection of original files used in this use case can be found in the NIRD research data archive (DOI: 10.11582/2021.00005). Note that the YAML specifications in the original dataset were compatible with immuneML version 0.0.91. This documentation page contains the YAML specifications for equivalent analyses with the latest immuneML version (last tested with version 1.1.3).
Generating synthetic immune repertoires with OLGA
We start by generating 2000 repertoires each containing 100000 amino acid sequences using OLGA. When OLGA is installed (see: OLGA GitHub page), the following piece of bash code can be used for this purpose:
for i in {1..2000}
do
echo "###### Repertoire $i"
olga-generate_sequences --humanIGH -o rep_$i.tsv -n 1e5 --seed=$i
done
Next, we can make a metadata file referring to all these repertoires using this piece of bash code:
echo "filename,subject_id" > metadata.csv
for i in {1..2000}
do
echo "rep_$i.tsv,$i" >> metadata.csv
done
Simulating immune signals into the immune repertoires
Next, five different immune signals are implanted in the 2000 immune repertoires. To read about immune signal simulation in more detail, see How to simulate antigen or disease-associated signals in AIRR datasets.
The implanted immune signals are of increasing complexity:
label 1: consists of one 3-mer,
label 2: consists of 20 3-mers
label 3: consists of 20 3-mers with gaps. The gap is alternating between positions
X/XXandXX/Xlabel 4: consists of 20 3-mers, but when implanting a 3-mer into a sequence, there is a 50% chance that one of its amino acids is randomly replaced with a different amino acid (hamming distance = 1)
label 5: consists of 20 3-mers combining both gaps and hamming distance as described for labels 3 and 4
The 3-mers for each label were randomly generated by choosing amino acids from a uniform distribution. Each label is implanted in half the repertoires, but the labels are not biased to occur together (each combination of two labels occurs in a quarter of the repertoires). When a label is implanted in a repertoire, it is implanted in 0.1% of the sequences. The kmers are implanted in a sequence starting at an IMGT position in the range [109, 112] (the middle of the CDR3 sequence), to make sure the conserved patterns near the beginning and end of the CDR3 sequence are not broken.
The following YAML specification represents how the immune signals were implanted in the 2000 repertoires, given that
the 2000 repertoires and metadata file generated in the previous step are located at path/to/olga_data/.
definitions:
datasets:
olga2000:
format: OLGA
params:
is_repertoire: true
path: path/to/olga_data/
metadata_file: path/to/olga_data/metadata.csv
motifs:
label1_motif1:
instantiation: GappedKmer
seed: YYG
label2_motif1:
instantiation: GappedKmer
seed: RRS
label2_motif10:
instantiation: GappedKmer
seed: NHE
label2_motif11:
instantiation: GappedKmer
seed: VDY
label2_motif12:
instantiation: GappedKmer
seed: KFA
label2_motif13:
instantiation: GappedKmer
seed: VLR
label2_motif14:
instantiation: GappedKmer
seed: ILT
label2_motif15:
instantiation: GappedKmer
seed: RIT
label2_motif16:
instantiation: GappedKmer
seed: CYT
label2_motif17:
instantiation: GappedKmer
seed: YII
label2_motif18:
instantiation: GappedKmer
seed: WLL
label2_motif19:
instantiation: GappedKmer
seed: FQP
label2_motif2:
instantiation: GappedKmer
seed: NQV
label2_motif20:
instantiation: GappedKmer
seed: YLG
label2_motif3:
instantiation: GappedKmer
seed: GYD
label2_motif4:
instantiation: GappedKmer
seed: RIH
label2_motif5:
instantiation: GappedKmer
seed: QHY
label2_motif6:
instantiation: GappedKmer
seed: FSR
label2_motif7:
instantiation: GappedKmer
seed: EGS
label2_motif8:
instantiation: GappedKmer
seed: YVS
label2_motif9:
instantiation: GappedKmer
seed: CRC
label3_motif1:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: RN/E
label3_motif10:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: M/NA
label3_motif11:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: TS/Y
label3_motif12:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: Y/YV
label3_motif13:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: PP/K
label3_motif14:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: D/ME
label3_motif15:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: SY/P
label3_motif16:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: V/NI
label3_motif17:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: YA/P
label3_motif18:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: E/KT
label3_motif19:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: MY/R
label3_motif2:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: D/IW
label3_motif20:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: N/DT
label3_motif3:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: IV/V
label3_motif4:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: T/CT
label3_motif5:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: EF/C
label3_motif6:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: N/IV
label3_motif7:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: RE/Q
label3_motif8:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: I/SM
label3_motif9:
instantiation:
GappedKmer:
max_gap: 1
min_gap: 0
seed: RD/H
label4_motif1:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: FQA
label4_motif10:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: RVY
label4_motif11:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: LPH
label4_motif12:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: PVW
label4_motif13:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: PSI
label4_motif14:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: FND
label4_motif15:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: WRP
label4_motif16:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: SVP
label4_motif17:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: LDV
label4_motif18:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: QTR
label4_motif19:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: MYN
label4_motif2:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: ASF
label4_motif20:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: HFR
label4_motif3:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: VPA
label4_motif4:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: DHE
label4_motif5:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: KTT
label4_motif6:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: RKG
label4_motif7:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: QIA
label4_motif8:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: RND
label4_motif9:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
seed: YWI
label5_motif1:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: RK/Q
label5_motif10:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: M/AF
label5_motif11:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: TY/C
label5_motif12:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: R/TA
label5_motif13:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: PV/G
label5_motif14:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: I/MR
label5_motif15:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: FT/R
label5_motif16:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: N/YV
label5_motif17:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: PH/W
label5_motif18:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: M/KC
label5_motif19:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: QL/S
label5_motif2:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: M/NS
label5_motif20:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: E/VI
label5_motif3:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: LR/N
label5_motif4:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: V/HM
label5_motif5:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: TV/V
label5_motif6:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: S/PD
label5_motif7:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: IW/M
label5_motif8:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: P/QN
label5_motif9:
instantiation:
GappedKmer:
hamming_distance_probabilities:
0: 0.5
1: 0.5
max_gap: 1
min_gap: 0
seed: YK/R
signals:
label1:
implanting: HealthySequence
motifs:
- label1_motif1
sequence_position_weights:
109: 1
110: 1
111: 1
112: 1
label2:
implanting: HealthySequence
motifs:
- label2_motif1
- label2_motif2
- label2_motif3
- label2_motif4
- label2_motif5
- label2_motif6
- label2_motif7
- label2_motif8
- label2_motif9
- label2_motif10
- label2_motif11
- label2_motif12
- label2_motif13
- label2_motif14
- label2_motif15
- label2_motif16
- label2_motif17
- label2_motif18
- label2_motif19
- label2_motif20
sequence_position_weights:
109: 1
110: 1
111: 1
112: 1
label3:
implanting: HealthySequence
motifs:
- label3_motif1
- label3_motif2
- label3_motif3
- label3_motif4
- label3_motif5
- label3_motif6
- label3_motif7
- label3_motif8
- label3_motif9
- label3_motif10
- label3_motif11
- label3_motif12
- label3_motif13
- label3_motif14
- label3_motif15
- label3_motif16
- label3_motif17
- label3_motif18
- label3_motif19
- label3_motif20
sequence_position_weights:
109: 1
110: 1
111: 1
112: 1
label4:
implanting: HealthySequence
motifs:
- label4_motif1
- label4_motif2
- label4_motif3
- label4_motif4
- label4_motif5
- label4_motif6
- label4_motif7
- label4_motif8
- label4_motif9
- label4_motif10
- label4_motif11
- label4_motif12
- label4_motif13
- label4_motif14
- label4_motif15
- label4_motif16
- label4_motif17
- label4_motif18
- label4_motif19
- label4_motif20
sequence_position_weights:
109: 1
110: 1
111: 1
112: 1
label5:
implanting: HealthySequence
motifs:
- label5_motif1
- label5_motif2
- label5_motif3
- label5_motif4
- label5_motif5
- label5_motif6
- label5_motif7
- label5_motif8
- label5_motif9
- label5_motif10
- label5_motif11
- label5_motif12
- label5_motif13
- label5_motif14
- label5_motif15
- label5_motif16
- label5_motif17
- label5_motif18
- label5_motif19
- label5_motif20
sequence_position_weights:
109: 1
110: 1
111: 1
112: 1
simulations:
sim1:
i1:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label1
- label2
- label3
i2:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label1
- label2
- label5
i3:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label1
- label3
- label4
- label5
i4:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label1
- label4
i5:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label2
- label3
- label4
i6:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label2
- label4
- label5
i7:
dataset_implanting_rate: 0.125
repertoire_implanting_rate: 0.001
signals:
- label3
- label5
instructions:
inst1:
dataset: olga2000
export_formats:
- AIRR
- ImmuneML
simulation: sim1
type: Simulation
output:
format: HTML
Benchmarking ML methods and encodings
Finally, we use the above-generated dataset with implanted disease signals for a benchmarking. We benchmark three different shallow ML methods: logistic regression, support vector machines and random forest. Each of these ML methods is combined with k-mer frequency encodings based on 3-mers and 4-mers. Because we use a ground truth benchmarking dataset where the true implanted signals are known, we use the MotifSeedRecovery report to show how well the k-mers recovered by the ML methods overlap with the k-mers that we originally implanted.
The YAML specification below shows the settings that were used for the benchmarking. We assume that the dataset
with simulated signals can be found at path/to/simulated_data/olga2000.iml_dataset.
Alternatively, you may want to use the AIRR files (airr.zip) that were produced in the original use case, which can be downloaded
from the NIRD research data archive (DOI: 10.11582/2021.00005).
In this case, uncomment the lines for AIRR import and remove the lines for ImmuneML import.
definitions:
datasets:
d1:
format: ImmuneML
params:
path: path/to/simulated_data/olga2000.iml_dataset
#d1:
#format: AIRR
#params:
#is_repertoire: true
#path: path/to/airr/repertoires/
#metadata_file: path/to/airr/metadata.csv
encodings:
3mer:
KmerFrequency:
k: 3
scale_to_unit_variance: true
scale_to_zero_mean: true
4mer:
KmerFrequency:
k: 4
scale_to_unit_variance: true
scale_to_zero_mean: true
ml_methods:
LR:
LogisticRegression:
C:
- 0.01
- 0.1
- 1
- 10
- 100
class_weight:
- balanced
max_iter:
- 1000
penalty:
- l1
model_selection_cv: true
model_selection_n_folds: 3
RF:
RandomForestClassifier:
n_estimators:
- 5
- 10
- 50
- 100
model_selection_cv: true
model_selection_n_folds: 3
SVM:
SVC:
C:
- 0.01
- 0.1
- 1
- 10
- 100
class_weight:
- balanced
max_iter:
- 1000
penalty:
- l1
dual: False
model_selection_cv: true
model_selection_n_folds: 3
reports:
coefs:
Coefficients:
coefs_to_plot:
- n_largest
n_largest:
- 25
name: coefs
hp_report:
MLSettingsPerformance:
name: hp_report
single_axis_labels: False
seeds:
MotifSeedRecovery:
gap_sizes:
- 1
hamming_distance: false
implanted_motifs_per_label:
signal_label1:
gap_sizes:
- 0
hamming_distance: false
seeds:
- YYG
signal_label2:
gap_sizes:
- 0
hamming_distance: false
seeds:
- RRS
- NHE
- VDY
- KFA
- VLR
- ILT
- RIT
- CYT
- YII
- WLL
- FQP
- NQV
- YLG
- GYD
- RIH
- QHY
- FSR
- EGS
- YVS
- CRC
signal_label3:
gap_sizes:
- 0
- 1
hamming_distance: false
seeds:
- RN/E
- M/NA
- TS/Y
- Y/YV
- PP/K
- D/ME
- SY/P
- V/NI
- YA/P
- E/KT
- MY/R
- D/IW
- N/DT
- IV/V
- T/CT
- EF/C
- N/IV
- RE/Q
- I/SM
- RD/H
signal_label4:
gap_sizes:
- 0
hamming_distance: true
seeds:
- FQA
- RVY
- LPH
- PVW
- PSI
- FND
- WRP
- SVP
- LDV
- QTR
- MYN
- ASF
- HFR
- VPA
- DHE
- KTT
- RKG
- QIA
- RND
- YWI
signal_label5:
gap_sizes:
- 0
- 1
hamming_distance: true
seeds:
- RK/Q
- M/AF
- TY/C
- R/TA
- PV/G
- I/MR
- FT/R
- N/YV
- PH/W
- QL/S
- M/NS
- E/VI
- LR/N
- V/HM
- TV/V
- S/PD
- IW/M
- P/QN
- YK/R
name: seeds
instructions:
inst1:
dataset: d1
labels:
- signal_label1
- signal_label2
- signal_label3
- signal_label4
- signal_label5
assessment:
reports:
models:
- coefs
- seeds
split_count: 3
split_strategy: random
training_percentage: 0.7
selection:
split_count: 1
split_strategy: random
training_percentage: 0.7
metrics:
- accuracy
- balanced_accuracy
optimization_metric: balanced_accuracy
settings:
- encoding: 3mer
ml_method: SVM
- encoding: 3mer
ml_method: LR
- encoding: 3mer
ml_method: RF
- encoding: 4mer
ml_method: SVM
- encoding: 4mer
ml_method: LR
- encoding: 4mer
ml_method: RF
reports:
- hp_report
refit_optimal_model: false
number_of_processes: 32
strategy: GridSearch
type: TrainMLModel
output:
format: HTML
Results
When benchmarking the three ML methods (logistic regression (LR), support vector machine (SVM) and random forest (RF)) in combination with two encodings (3-mer and 4-mer encoding) using the synthetic datset with ground-truth disease signals, we show that the classification performance drops as the immune event complexity increases:
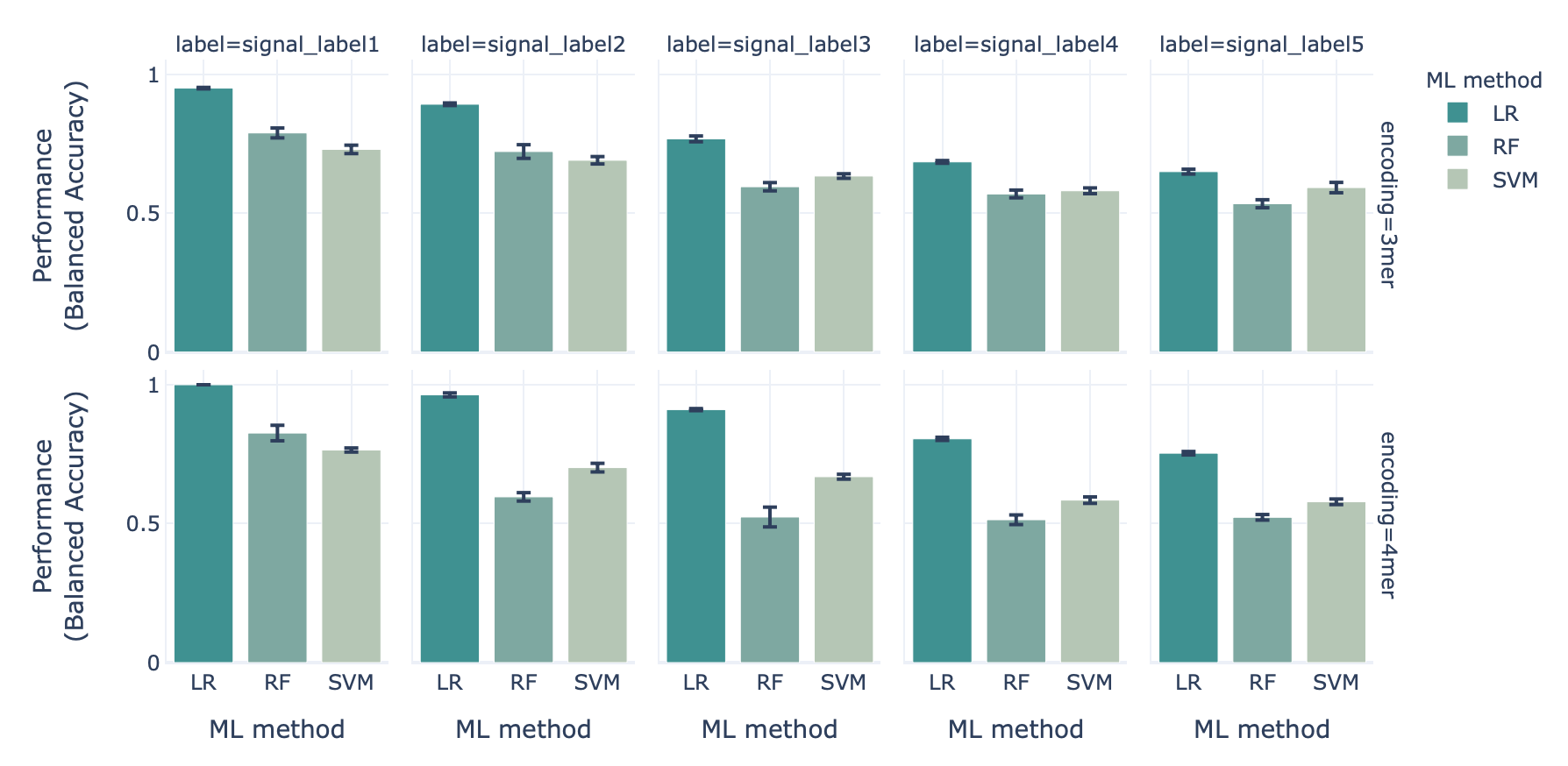
The classification performance for the most simple immune signal (signal 1) is highest, while for the most complex immune signal (signal 5) it is lowest.
Furthermore, when comparing the feature coefficient sizes with how well these features represent the ground-truth signals, it was found that models with a good classification performance were indeed able to recover the ground-truth signals (here only shown for immune signals 1 and 5, for data split 1).
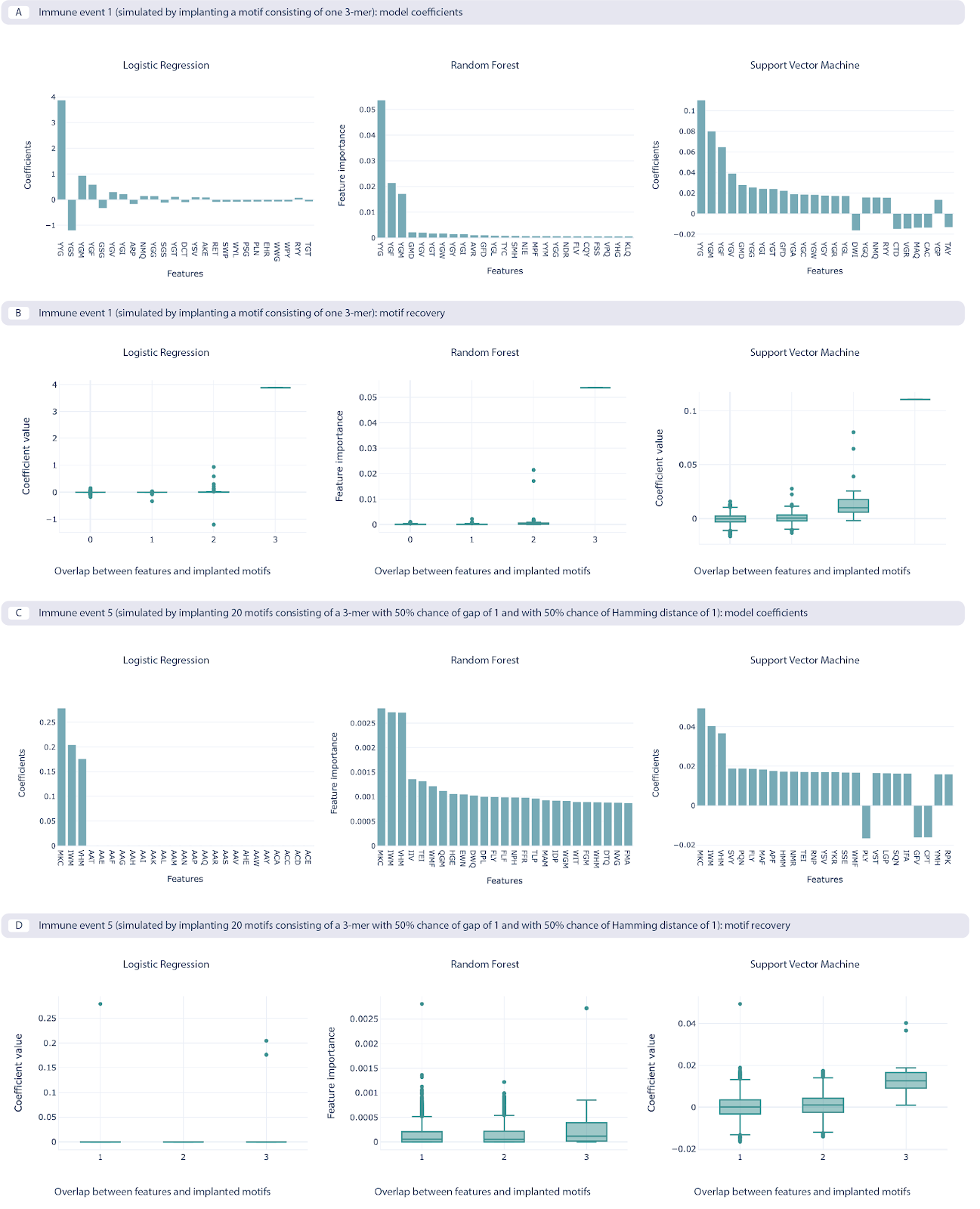
The benchmarking use case model coefficients and motif recovery, where the repertoire data is represented by 3-mer amino acid frequencies. Two immune events are shown. Immune event 1 (A, B) is the simplest event simulated by implanting a single 3-mer, while the immune event 5 (C, D) is the most complex one simulated by implanting 20 motifs consisting of a 3-mer with a 50% chance of having a gap and 50% chance of having a Hamming distance of 1. A. The 25 largest coefficients of the logistic regression model, feature importances on random forest model, and coefficients of the support vector machine (SVM) model. The highest value of the coefficients corresponds to the implanted motif. B. Coefficient values for the features depending on the overlap between the recovered features that overlap with the implanted motif, measuring how well the recovered motifs correspond to the implanted motif, shown across the three ML models. C. The 25 largest coefficients and feature importances for the ML models trained on immune event 5. D. Overlap of recovered and implanted motifs for the ML models trained on immune event 5. Motif recovery for immune event 5 is less effective than for immune event 1.