immuneML.ml_methods package
Submodules
immuneML.ml_methods.AtchleyKmerMILClassifier module
- class immuneML.ml_methods.AtchleyKmerMILClassifier.AtchleyKmerMILClassifier(iteration_count: Optional[int] = None, threshold: Optional[float] = None, evaluate_at: Optional[int] = None, use_early_stopping: Optional[bool] = None, random_seed: Optional[int] = None, learning_rate: Optional[float] = None, zero_abundance_weight_init: Optional[bool] = None, number_of_threads: Optional[int] = None, result_path: Optional[pathlib.Path] = None)[source]
Bases:
immuneML.ml_methods.MLMethod.MLMethodA binary Repertoire classifier which uses the data encoded by AtchleyKmer encoder to predict the repertoire label.
The original publication: Ostmeyer J, Christley S, Toby IT, Cowell LG. Biophysicochemical motifs in T cell receptor sequences distinguish repertoires from tumor-infiltrating lymphocytes and adjacent healthy tissue. Cancer Res. Published online January 1, 2019:canres.2292.2018. doi:10.1158/0008-5472.CAN-18-2292 .
- Parameters
iteration_count (int) – max number of training iterations
threshold (float) – loss threshold at which to stop training if reached
evaluate_at (int) – log model performance every ‘evaluate_at’ iterations and store the model every ‘evaluate_at’ iterations if early stopping
used (is) –
use_early_stopping (bool) – whether to use early stopping
learning_rate (float) – learning rate for stochastic gradient descent
random_seed (int) – random seed used
zero_abundance_weight_init (bool) – whether to use 0 as initial weight for abundance term (if not, a random value is sampled from normal
total_number_of_features (distribution with mean 0 and variance 1 /) –
number_of_threads – number of threads to be used for training
YAML specification:
my_kmer_mil_classifier: AtchleyKmerMILClassifier: iteration_count: 100 evaluate_at: 15 use_early_stopping: False learning_rate: 0.01 random_seed: 100 zero_abundance_weight_init: True number_of_threads: 8 threshold: 0.00001
- can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- check_if_exists(path) bool[source]
The check_if_exists function checks if there is a stored model on the given path. Might be useful in the future for implementing checkpoints. See SklearnMethod for example usage.
- Parameters
path (Path) – path to folder where it should be checked if the model was stored previously
- Returns
whether the model was stored previously on the given path or not
- Return type
True/False
- fit(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label, cores_for_training: int = 2)[source]
The fit function fits the parameters of the machine learning model.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – if parallelization is available in the MLMethod (and the availability depends on the specific classifier), this is the number of processes that will be creating when fitting the model to speed up the computation.
- Returns
it doesn’t return anything, but fits the model parameters instead
- fit_by_cross_validation(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, number_of_splits: int = 5, label: Optional[immuneML.environment.Label.Label] = None, cores_for_training: int = - 1, optimization_metric=None)[source]
The fit_by_cross_validation function should implement finding the best model hyperparameters through cross-validation. In immuneML, preprocessing, encoding and ML hyperparameters can be optimized by using nested cross-validation (see TrainMLModelInstruction for more details). This function is in that setting the third level of nested cross-validation as it can optimize only over the model hyperparameters. It represents an alternative to optimizing the model hyperparameters in the TrainMLModelInstruction. Which one should be used depends on the use-case and specific models: models based on scikit-learn implementations come with this option by default (see SklearnMethod class), while custom classifiers typically do not implement this and just call fit() function and throw a warning instead.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
number_of_splits (int) – number of splits for the cross-validation to be performed for selection the best hyperparameters of the ML model; note that if this is used in combination with nested cross-validation in TrainMLModel instruction, it can result in very few examples in each split depending on the orginal dataset size and the nested cross-validation setup.
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – number of processes to be used during the cross-validation for model selection
optimization_metric (str) – the name of the optimization metric to be used to select the best model during cross-validation; when used with TrainMLModel instruction which is almost exclusively the case when the immuneML is run from the specification, this maps to the optimization metric in the instruction.
- Returns
it doesn’t return anything, but fits the model parameters instead
- get_class_mapping() dict[source]
Returns a dictionary containing the mapping between label values and values internally used in the classifier
- get_feature_names() list[source]
Returns the list of feature names (a list of strings) if available where the feature names were provided by the encoder in the EncodedData object.
- get_package_info() str[source]
Returns the package and version used for implementing the ML method if an external package was used or immuneML version if it is custom implementation. See py:mod:immuneML.ml_methods.SklearnMethod.SklearnMethod and py:mod:immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier for examples of both.
- get_params()[source]
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- load(path)[source]
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters
path (Path) – path to the folder where the model was stored using store() function
- Returns
it does not have a return value, but sets the attribute values of the object instead
- predict(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict function predicts the class for the given label across examples provided in encoded data.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label – (Label): the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
- Returns
e.g., {label_name: [class1, class2, class2, class1]}
- Return type
a dictionary where the key is the label_name and the value is a list of class predictions (one prediction per example)
- predict_proba(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict_proba function predicts class probabilities for the given label if the model supports probabilistic output. If not, it should raise a warning and return predicted classes without probabilities.
Note that when providing class probabilities the classes should have a specific (constant) order, and in case of binary classification, they should be ordered so that the negative class comes first and the positive one comes second. For this handling classes, see py:mod:immuneML.ml_methods.util.Util.Util.make_binary_class_mapping method that will automatically create class mapping for binary classification.
- Parameters
encoded_data (EncodedData) – an object of EncodedData class where the examples attribute should be used to make predictions. examples
format (attribute includes encoded examples in matrix) –
labels (provided here can include) –
labels) (been) –
function (so the labels attribute of the EncodedData object should NOT be used in this) –
set. (even if it is) –
label (Label) – the label for which the prediction should be made. It can be used to check if it matches the label that the
not (model has been trained for and if) –
any (an exception should be thrown. It is often an AssertionError as this can be checked before) –
made (prediction is) –
cases (but could also be a RuntimeError. It both) –
message. (it should include a user-friendly) –
- Returns
a dictionary where the key is the label name and the value a 2D numpy array with class probabilities of dimension [number_of_examples x number_of_classes_for_label], for instance for label CMV where the class can be either True or False and there are 3 examples to predict the class probabilities for: {CMV: [[0.2, 0.8], [0.55, 0.45], [0.98, 0.02]]}
- store(path: pathlib.Path, feature_names=None, details_path: Optional[pathlib.Path] = None)[source]
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters
path (Path) – path to folder where to store the model
feature_names (list) – list of feature names in the encoded data; this can be stored as well to make it easier to map linear models to specific features as provided by the encoded (e.g., in case of logistic regression, this feature list defines what coefficients refer to)
details_path (Path) – path to folder where to store the details of the model. The details can be there to better understand the model but are not mandatory and are typically not loaded with the model afterwards. This is user-friendly file that can be examined manually by the user. It does not have to be created or can be created at the same folder as the path parameters points to. In practice, when used with TrainMLModel instruction, this parameter will either be None or have the same value as path parameter.
- Returns
it does not have a return value
immuneML.ml_methods.DeepRC module
- class immuneML.ml_methods.DeepRC.DeepRC(validation_part, add_positional_information, kernel_size, n_kernels, n_additional_convs, n_attention_network_layers, n_attention_network_units, n_output_network_units, consider_seq_counts, sequence_reduction_fraction, reduction_mb_size, n_updates, n_torch_threads, learning_rate, l1_weight_decay, l2_weight_decay, evaluate_at, sample_n_sequences, training_batch_size, n_workers, keep_dataset_in_ram, pytorch_device_name)[source]
Bases:
immuneML.ml_methods.MLMethod.MLMethodThis classifier uses the DeepRC method for repertoire classification. The DeepRC ML method should be used in combination with the DeepRC encoder. Also consider using the DeepRCMotifDiscovery report for interpretability.
Notes:
DeepRC uses PyTorch functionalities that depend on GPU. Therefore, DeepRC does not work on a CPU.
This wrapper around DeepRC currently only supports binary classification.
Reference: Michael Widrich, Bernhard Schäfl, Milena Pavlović, Geir Kjetil Sandve, Sepp Hochreiter, Victor Greiff, Günter Klambauer ‘DeepRC: Immune repertoire classification with attention-based deep massive multiple instance learning’. bioRxiv preprint doi: https://doi.org/10.1101/2020.04.12.038158
- Parameters
validation_part (float) – the part of the data that will be used for validation, the rest will be used for training.
add_positional_information (bool) – whether positional information should be included in the input features.
kernel_size (int) – the size of the 1D-CNN kernels.
n_kernels (int) – the number of 1D-CNN kernels in each layer.
n_additional_convs (int) – Number of additional 1D-CNN layers after first layer
n_attention_network_layers (int) – Number of attention layers to compute keys
n_attention_network_units (int) – Number of units in each attention layer
n_output_network_units (int) – Number of units in the output layer
consider_seq_counts (bool) – whether the input data should be scaled by the receptor sequence counts.
sequence_reduction_fraction (float) – Fraction of number of sequences to which to reduce the number of sequences per bag based on attention weights. Has to be in range [0,1].
reduction_mb_size (int) – Reduction of sequences per bag is performed using minibatches of reduction_mb_size` sequences to compute the attention weights.
n_updates (int) – Number of updates to train for
n_torch_threads (int) – Number of parallel threads to allow PyTorch
learning_rate (float) – Learning rate for adam optimizer
l1_weight_decay (float) – l1 weight decay factor. l1 weight penalty will be added to loss, scaled by l1_weight_decay
l2_weight_decay (float) – l2 weight decay factor. l2 weight penalty will be added to loss, scaled by l2_weight_decay
evaluate_at (int) – Evaluate model on training and validation set every evaluate_at updates. This will also check for a new best model for early stopping.
sample_n_sequences (int) – Optional random sub-sampling of sample_n_sequences sequences per repertoire. Number of sequences per repertoire might be smaller than sample_n_sequences if repertoire is smaller or random indices have been drawn multiple times. If None, all sequences will be loaded for each repertoire.
training_batch_size (int) – Number of repertoires per minibatch during training.
n_workers (int) – Number of background processes to use for converting dataset to hdf5 container and training set data loader.
pytorch_device_name (str) – The name of the pytorch device to use. This name will be passed to torch.device(self.pytorch_device_name). The default value is cuda:0
YAML specification:
my_deeprc_method: DeepRC: validation_part: 0.2 add_positional_information: True kernel_size: 9
- can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- check_if_exists(path)[source]
The check_if_exists function checks if there is a stored model on the given path. Might be useful in the future for implementing checkpoints. See SklearnMethod for example usage.
- Parameters
path (Path) – path to folder where it should be checked if the model was stored previously
- Returns
whether the model was stored previously on the given path or not
- Return type
True/False
- fit(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label, cores_for_training: int = 2)[source]
The fit function fits the parameters of the machine learning model.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – if parallelization is available in the MLMethod (and the availability depends on the specific classifier), this is the number of processes that will be creating when fitting the model to speed up the computation.
- Returns
it doesn’t return anything, but fits the model parameters instead
- fit_by_cross_validation(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, number_of_splits: int = 5, label: Optional[immuneML.environment.Label.Label] = None, cores_for_training: int = - 1, optimization_metric=None)[source]
The fit_by_cross_validation function should implement finding the best model hyperparameters through cross-validation. In immuneML, preprocessing, encoding and ML hyperparameters can be optimized by using nested cross-validation (see TrainMLModelInstruction for more details). This function is in that setting the third level of nested cross-validation as it can optimize only over the model hyperparameters. It represents an alternative to optimizing the model hyperparameters in the TrainMLModelInstruction. Which one should be used depends on the use-case and specific models: models based on scikit-learn implementations come with this option by default (see SklearnMethod class), while custom classifiers typically do not implement this and just call fit() function and throw a warning instead.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
number_of_splits (int) – number of splits for the cross-validation to be performed for selection the best hyperparameters of the ML model; note that if this is used in combination with nested cross-validation in TrainMLModel instruction, it can result in very few examples in each split depending on the orginal dataset size and the nested cross-validation setup.
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – number of processes to be used during the cross-validation for model selection
optimization_metric (str) – the name of the optimization metric to be used to select the best model during cross-validation; when used with TrainMLModel instruction which is almost exclusively the case when the immuneML is run from the specification, this maps to the optimization metric in the instruction.
- Returns
it doesn’t return anything, but fits the model parameters instead
- get_class_mapping() dict[source]
Returns a dictionary containing the mapping between label values and values internally used in the classifier
- get_feature_names() list[source]
Returns the list of feature names (a list of strings) if available where the feature names were provided by the encoder in the EncodedData object.
- get_package_info() str[source]
Returns the package and version used for implementing the ML method if an external package was used or immuneML version if it is custom implementation. See py:mod:immuneML.ml_methods.SklearnMethod.SklearnMethod and py:mod:immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier for examples of both.
- get_params()[source]
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- load(path: pathlib.Path, details_path: Optional[pathlib.Path] = None)[source]
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters
path (Path) – path to the folder where the model was stored using store() function
- Returns
it does not have a return value, but sets the attribute values of the object instead
- make_data_loader(hdf5_filepath: pathlib.Path, pre_loaded_hdf5_file, indices, label_name, eval_only: bool, is_train: bool, n_workers=1)[source]
Creates a pytorch dataloader using DeepRC’s RepertoireDataReaderBinary
- Parameters
hdf5_filepath – the path to the HDF5 file
pre_loaded_hdf5_file – Optional: It is faster to load the hdf5 file into the RAM as dictionary instead of keeping it on the disk. pre_loaded_hdf5_file is the loaded hdf5 file as dictionary. If None, the hdf5 file will be read from the disk and consume less RAM.
indices – indices of the subset of repertoires in the data that will be used for this dataset. If ‘None’, all repertoires will be used.
label_name – the name of the label to be predicted
eval_only – whether the dataloader will only be used for evaluation (no training). if false, sample_n_sequences can be set
is_train – whether this is a dataloader for training data. If true, self.training_batch_size is used.
n_workers – the number of workers used in torch.utils.data.DataLoader
- Returns
a Pytorch dataloader
- predict(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict function predicts the class for the given label across examples provided in encoded data.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label – (Label): the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
- Returns
e.g., {label_name: [class1, class2, class2, class1]}
- Return type
a dictionary where the key is the label_name and the value is a list of class predictions (one prediction per example)
- predict_proba(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict_proba function predicts class probabilities for the given label if the model supports probabilistic output. If not, it should raise a warning and return predicted classes without probabilities.
Note that when providing class probabilities the classes should have a specific (constant) order, and in case of binary classification, they should be ordered so that the negative class comes first and the positive one comes second. For this handling classes, see py:mod:immuneML.ml_methods.util.Util.Util.make_binary_class_mapping method that will automatically create class mapping for binary classification.
- Parameters
encoded_data (EncodedData) – an object of EncodedData class where the examples attribute should be used to make predictions. examples
format (attribute includes encoded examples in matrix) –
labels (provided here can include) –
labels) (been) –
function (so the labels attribute of the EncodedData object should NOT be used in this) –
set. (even if it is) –
label (Label) – the label for which the prediction should be made. It can be used to check if it matches the label that the
not (model has been trained for and if) –
any (an exception should be thrown. It is often an AssertionError as this can be checked before) –
made (prediction is) –
cases (but could also be a RuntimeError. It both) –
message. (it should include a user-friendly) –
- Returns
a dictionary where the key is the label name and the value a 2D numpy array with class probabilities of dimension [number_of_examples x number_of_classes_for_label], for instance for label CMV where the class can be either True or False and there are 3 examples to predict the class probabilities for: {CMV: [[0.2, 0.8], [0.55, 0.45], [0.98, 0.02]]}
- store(path, feature_names=None, details_path: Optional[pathlib.Path] = None)[source]
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters
path (Path) – path to folder where to store the model
feature_names (list) – list of feature names in the encoded data; this can be stored as well to make it easier to map linear models to specific features as provided by the encoded (e.g., in case of logistic regression, this feature list defines what coefficients refer to)
details_path (Path) – path to folder where to store the details of the model. The details can be there to better understand the model but are not mandatory and are typically not loaded with the model afterwards. This is user-friendly file that can be examined manually by the user. It does not have to be created or can be created at the same folder as the path parameters points to. In practice, when used with TrainMLModel instruction, this parameter will either be None or have the same value as path parameter.
- Returns
it does not have a return value
immuneML.ml_methods.KNN module
- class immuneML.ml_methods.KNN.KNN(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodThis is a wrapper of scikit-learn’s KNeighborsClassifier class. This ML method creates a distance matrix using the given encoded data. If the encoded data is already a distance matrix (for example, when using the Distance or CompAIRRDistance encoders), please use PrecomputedKNN instead.
Please see the scikit-learn documentation of KNeighborsClassifier for the parameters.
For usage instructions, check
SklearnMethod.YAML specification:
my_knn_method: KNN: # sklearn parameters (same names as in original sklearn class) weights: uniform # always use this setting for weights n_neighbors: [5, 10, 15] # find the optimal number of neighbors # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under KNN is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_knn: KNN
immuneML.ml_methods.LogisticRegression module
- class immuneML.ml_methods.LogisticRegression.LogisticRegression(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodThis is a wrapper of scikit-learn’s LogisticRegression class. Please see the scikit-learn documentation of LogisticRegression for the parameters.
Note: if you are interested in plotting the coefficients of the logistic regression model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
my_logistic_regression: # user-defined method name LogisticRegression: # name of the ML method # sklearn parameters (same names as in original sklearn class) penalty: l1 # always use penalty l1 C: [0.01, 0.1, 1, 10, 100] # find the optimal value for C # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under LogisticRegression is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_logistic_regression: LogisticRegression
- can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- default_parameters = {'max_iter': 1000, 'solver': 'saga'}
immuneML.ml_methods.MLMethod module
- class immuneML.ml_methods.MLMethod.MLMethod[source]
Bases:
objectBase class for different machine learning methods, defining which functions should be implemented. These public functions are the only ones that will be used outside the method, during training, assessment or while making predictions. Most often the methods will be classifiers (binary or multi-class) that should learn some label on either immune repertoires (sets of receptor sequences), receptors (paired sequences) or receptor sequences (lists of amino acids).
Here we refer to machine learning methods (algorithms) as a method that, given a set of examples and corresponding labels, constructs a model (such as logistic regression), whereas we define the model to be already fit to data using the learning method (algorithm), such as logistic regression with specific coefficients.
The functions of this class provide a standard set of ML functions: fitting the model (with or without cross-validation) and making predictions (either class predictions or class probabilities if possible). Other functions provide for various utilities, such as storing and loading the model, checking if it was fit already, retrieving coefficients for user-friendly output etc.
Note that when providing class probabilities the classes should have a specific (constant) order, and in case of binary classification, they should be ordered so that the negative class comes first and the positive one comes second. For this handling classes, see py:immuneML.ml_methods.util.Util.Util.make_class_mapping method that will automatically create class mapping for classification.
- abstract can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- check_encoder_compatibility(encoder)[source]
Checks whether the given encoder is compatible with this ML method, and throws an error if it is not.
- abstract check_if_exists(path: pathlib.Path) bool[source]
The check_if_exists function checks if there is a stored model on the given path. Might be useful in the future for implementing checkpoints. See SklearnMethod for example usage.
- Parameters
path (Path) – path to folder where it should be checked if the model was stored previously
- Returns
whether the model was stored previously on the given path or not
- Return type
True/False
- abstract fit(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: <module 'immuneML.environment.Label' from '/Users/milenpa/PycharmProjects/BMIImmuneML/immuneML/environment/Label.py'>, cores_for_training: int = 2)[source]
The fit function fits the parameters of the machine learning model.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – if parallelization is available in the MLMethod (and the availability depends on the specific classifier), this is the number of processes that will be creating when fitting the model to speed up the computation.
- Returns
it doesn’t return anything, but fits the model parameters instead
- abstract fit_by_cross_validation(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, number_of_splits: int = 5, label: <module 'immuneML.environment.Label' from '/Users/milenpa/PycharmProjects/BMIImmuneML/immuneML/environment/Label.py'> = None, cores_for_training: int = -1, optimization_metric=None)[source]
The fit_by_cross_validation function should implement finding the best model hyperparameters through cross-validation. In immuneML, preprocessing, encoding and ML hyperparameters can be optimized by using nested cross-validation (see TrainMLModelInstruction for more details). This function is in that setting the third level of nested cross-validation as it can optimize only over the model hyperparameters. It represents an alternative to optimizing the model hyperparameters in the TrainMLModelInstruction. Which one should be used depends on the use-case and specific models: models based on scikit-learn implementations come with this option by default (see SklearnMethod class), while custom classifiers typically do not implement this and just call fit() function and throw a warning instead.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
number_of_splits (int) – number of splits for the cross-validation to be performed for selection the best hyperparameters of the ML model; note that if this is used in combination with nested cross-validation in TrainMLModel instruction, it can result in very few examples in each split depending on the orginal dataset size and the nested cross-validation setup.
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – number of processes to be used during the cross-validation for model selection
optimization_metric (str) – the name of the optimization metric to be used to select the best model during cross-validation; when used with TrainMLModel instruction which is almost exclusively the case when the immuneML is run from the specification, this maps to the optimization metric in the instruction.
- Returns
it doesn’t return anything, but fits the model parameters instead
- abstract get_class_mapping() dict[source]
Returns a dictionary containing the mapping between label values and values internally used in the classifier
- abstract get_feature_names() list[source]
Returns the list of feature names (a list of strings) if available where the feature names were provided by the encoder in the EncodedData object.
- abstract get_label_name() str[source]
Returns the name of the label for which the model was fitted.
- abstract get_package_info() str[source]
Returns the package and version used for implementing the ML method if an external package was used or immuneML version if it is custom implementation. See py:mod:immuneML.ml_methods.SklearnMethod.SklearnMethod and py:mod:immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier for examples of both.
- abstract get_params()[source]
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- abstract load(path: pathlib.Path)[source]
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters
path (Path) – path to the folder where the model was stored using store() function
- Returns
it does not have a return value, but sets the attribute values of the object instead
- abstract predict(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: <module 'immuneML.environment.Label' from '/Users/milenpa/PycharmProjects/BMIImmuneML/immuneML/environment/Label.py'>)[source]
The predict function predicts the class for the given label across examples provided in encoded data.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label – (Label): the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
- Returns
e.g., {label_name: [class1, class2, class2, class1]}
- Return type
a dictionary where the key is the label_name and the value is a list of class predictions (one prediction per example)
- abstract predict_proba(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, Label: <module 'immuneML.environment.Label' from '/Users/milenpa/PycharmProjects/BMIImmuneML/immuneML/environment/Label.py'>)[source]
The predict_proba function predicts class probabilities for the given label if the model supports probabilistic output. If not, it should raise a warning and return predicted classes without probabilities.
Note that when providing class probabilities the classes should have a specific (constant) order, and in case of binary classification, they should be ordered so that the negative class comes first and the positive one comes second. For this handling classes, see py:mod:immuneML.ml_methods.util.Util.Util.make_binary_class_mapping method that will automatically create class mapping for binary classification.
- Parameters
encoded_data (EncodedData) – an object of EncodedData class where the examples attribute should be used to make predictions. examples
format (attribute includes encoded examples in matrix) –
labels (provided here can include) –
labels) (been) –
function (so the labels attribute of the EncodedData object should NOT be used in this) –
set. (even if it is) –
label (Label) – the label for which the prediction should be made. It can be used to check if it matches the label that the
not (model has been trained for and if) –
any (an exception should be thrown. It is often an AssertionError as this can be checked before) –
made (prediction is) –
cases (but could also be a RuntimeError. It both) –
message. (it should include a user-friendly) –
- Returns
a dictionary where the key is the label name and the value a 2D numpy array with class probabilities of dimension [number_of_examples x number_of_classes_for_label], for instance for label CMV where the class can be either True or False and there are 3 examples to predict the class probabilities for: {CMV: [[0.2, 0.8], [0.55, 0.45], [0.98, 0.02]]}
- abstract store(path: pathlib.Path, feature_names: Optional[list] = None, details_path: Optional[pathlib.Path] = None)[source]
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters
path (Path) – path to folder where to store the model
feature_names (list) – list of feature names in the encoded data; this can be stored as well to make it easier to map linear models to specific features as provided by the encoded (e.g., in case of logistic regression, this feature list defines what coefficients refer to)
details_path (Path) – path to folder where to store the details of the model. The details can be there to better understand the model but are not mandatory and are typically not loaded with the model afterwards. This is user-friendly file that can be examined manually by the user. It does not have to be created or can be created at the same folder as the path parameters points to. In practice, when used with TrainMLModel instruction, this parameter will either be None or have the same value as path parameter.
- Returns
it does not have a return value
immuneML.ml_methods.PrecomputedKNN module
- class immuneML.ml_methods.PrecomputedKNN.PrecomputedKNN(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodThis is a wrapper of scikit-learn’s KNeighborsClassifier class. This ML method takes a pre-computed distance matrix, as created by the Distance or CompAIRRDistance encoders. If you would like to use a different encoding in combination with KNN, please use KNN instead.
Please see the scikit-learn documentation of KNeighborsClassifier for the parameters.
For usage instructions, check
SklearnMethod.YAML specification:
my_knn_method: KNN: # sklearn parameters (same names as in original sklearn class) weights: uniform # always use this setting for weights n_neighbors: [5, 10, 15] # find the optimal number of neighbors # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under KNN is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_knn: KNN
immuneML.ml_methods.ProbabilisticBinaryClassifier module
- class immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier(max_iterations: Optional[int] = None, update_rate: Optional[float] = None, likelihood_threshold: Optional[float] = None)[source]
Bases:
immuneML.ml_methods.MLMethod.MLMethodProbabilisticBinaryClassifier predicts the class assignment in binary classification case based on encoding examples by number of successful trials and total number of trials. It models this ratio by one beta distribution per class and predicts the class of the new examples using log-posterior odds ratio with threshold at 0.
ProbabilisticBinaryClassifier is based on the paper (details on the classification can be found in the Online Methods section): Emerson, Ryan O., William S. DeWitt, Marissa Vignali, Jenna Gravley, Joyce K. Hu, Edward J. Osborne, Cindy Desmarais, et al. ‘Immunosequencing Identifies Signatures of Cytomegalovirus Exposure History and HLA-Mediated Effects on the T Cell Repertoire’. Nature Genetics 49, no. 5 (May 2017): 659–65. doi.org/10.1038/ng.3822.
- Parameters
max_iterations (int) – maximum number of iterations while optimizing the parameters of the beta distribution (same for both classes)
update_rate (float) – how much the computed gradient should influence the updated value of the parameters of the beta distribution
likelihood_threshold (float) – at which threshold to stop the optimization (default -1e-10)
YAML specification:
my_probabilistic_classifier: # user-defined name of the ML method ProbabilisticBinaryClassifier: # method name max_iterations: 1000 update_rate: 0.01
- SMALL_POSITIVE_NUMBER = 1e-15
- can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- check_if_exists(path)[source]
The check_if_exists function checks if there is a stored model on the given path. Might be useful in the future for implementing checkpoints. See SklearnMethod for example usage.
- Parameters
path (Path) – path to folder where it should be checked if the model was stored previously
- Returns
whether the model was stored previously on the given path or not
- Return type
True/False
- fit(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label, cores_for_training: int = 2)[source]
The fit function fits the parameters of the machine learning model.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – if parallelization is available in the MLMethod (and the availability depends on the specific classifier), this is the number of processes that will be creating when fitting the model to speed up the computation.
- Returns
it doesn’t return anything, but fits the model parameters instead
- fit_by_cross_validation(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, number_of_splits: int = 5, label: Optional[immuneML.environment.Label.Label] = None, cores_for_training: int = - 1, optimization_metric=None)[source]
The fit_by_cross_validation function should implement finding the best model hyperparameters through cross-validation. In immuneML, preprocessing, encoding and ML hyperparameters can be optimized by using nested cross-validation (see TrainMLModelInstruction for more details). This function is in that setting the third level of nested cross-validation as it can optimize only over the model hyperparameters. It represents an alternative to optimizing the model hyperparameters in the TrainMLModelInstruction. Which one should be used depends on the use-case and specific models: models based on scikit-learn implementations come with this option by default (see SklearnMethod class), while custom classifiers typically do not implement this and just call fit() function and throw a warning instead.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
number_of_splits (int) – number of splits for the cross-validation to be performed for selection the best hyperparameters of the ML model; note that if this is used in combination with nested cross-validation in TrainMLModel instruction, it can result in very few examples in each split depending on the orginal dataset size and the nested cross-validation setup.
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – number of processes to be used during the cross-validation for model selection
optimization_metric (str) – the name of the optimization metric to be used to select the best model during cross-validation; when used with TrainMLModel instruction which is almost exclusively the case when the immuneML is run from the specification, this maps to the optimization metric in the instruction.
- Returns
it doesn’t return anything, but fits the model parameters instead
- get_class_mapping() dict[source]
Returns a dictionary containing the mapping between label values and values internally used in the classifier
- get_feature_names() list[source]
Returns the list of feature names (a list of strings) if available where the feature names were provided by the encoder in the EncodedData object.
- get_package_info() str[source]
Returns the package and version used for implementing the ML method if an external package was used or immuneML version if it is custom implementation. See py:mod:immuneML.ml_methods.SklearnMethod.SklearnMethod and py:mod:immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier for examples of both.
- get_params()[source]
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- load(path: pathlib.Path)[source]
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters
path (Path) – path to the folder where the model was stored using store() function
- Returns
it does not have a return value, but sets the attribute values of the object instead
- predict(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
Predict the class assignment for examples in X (where X is validation or test set - examples not seen during training).
\[\begin{split}\widehat{c} \, (k, n) = \left\{\begin{matrix} 0, & F(k, n) \leq 0\\ 1, & F(k, n) > 0 \end{matrix}\right\end{split}\]- Parameters
encoded_data (EncodedData) – EncodedData object with examples attribute which is a design matrix of shape
features] ([number of examples x number of) –
sequences (where number of features is 2 (the first feature is the number of disease-associated) –
example) (and the second is the total number of sequences per) –
label (str) – the label used for classification (e.g. CMV)
- Returns
{label_name: predictions} where predictions is a list of predicted classes for each example
- Return type
a dictionary of the following format
- predict_proba(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
Predict the probability of the class for examples in X.
\[\widehat{c} \, (k, n) = '\left\{\begin{matrix} 0, & F(k, n) \leq 0\ 1, & F(k, n) > 0 \end{matrix}\right\]- Parameters
encoded_data (EncodedData) – EncodedData object with examples attribute which is a design matrix of shape, where number of features is 2
example) ((the first feature is the number of disease-associated sequences and the second is the total number of sequences per) –
label (str) – the label used for classification (e.g. CMV)
- Returns
class probabilities for all examples in X
- store(path: pathlib.Path, feature_names=None, details_path=None)[source]
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters
path (Path) – path to folder where to store the model
feature_names (list) – list of feature names in the encoded data; this can be stored as well to make it easier to map linear models to specific features as provided by the encoded (e.g., in case of logistic regression, this feature list defines what coefficients refer to)
details_path (Path) – path to folder where to store the details of the model. The details can be there to better understand the model but are not mandatory and are typically not loaded with the model afterwards. This is user-friendly file that can be examined manually by the user. It does not have to be created or can be created at the same folder as the path parameters points to. In practice, when used with TrainMLModel instruction, this parameter will either be None or have the same value as path parameter.
- Returns
it does not have a return value
immuneML.ml_methods.RandomForestClassifier module
- class immuneML.ml_methods.RandomForestClassifier.RandomForestClassifier(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodThis is a wrapper of scikit-learn’s RandomForestClassifier class. Please see the scikit-learn documentation of RandomForestClassifier for the parameters.
Note: if you are interested in plotting the coefficients of the random forest classifier model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
my_random_forest_classifier: # user-defined method name RandomForestClassifier: # name of the ML method # sklearn parameters (same names as in original sklearn class) random_state: 100 # always use this value for random state n_estimators: [10, 50, 100] # find the optimal number of trees in the forest # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under RandomForestClassifier is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_random_forest: RandomForestClassifier
immuneML.ml_methods.ReceptorCNN module
- class immuneML.ml_methods.ReceptorCNN.ReceptorCNN(kernel_count: Optional[int] = None, kernel_size=None, positional_channels: Optional[int] = None, sequence_type: Optional[str] = None, device=None, number_of_threads: Optional[int] = None, random_seed: Optional[int] = None, learning_rate: Optional[float] = None, iteration_count: Optional[int] = None, l1_weight_decay: Optional[float] = None, l2_weight_decay: Optional[float] = None, batch_size: Optional[int] = None, training_percentage: Optional[float] = None, evaluate_at: Optional[int] = None, background_probabilities=None, result_path: Optional[pathlib.Path] = None)[source]
Bases:
immuneML.ml_methods.MLMethod.MLMethodA CNN which separately detects motifs using CNN kernels in each chain of paired receptor data, combines the kernel activations into a unique representation of the receptor and uses this representation to predict the antigen binding.
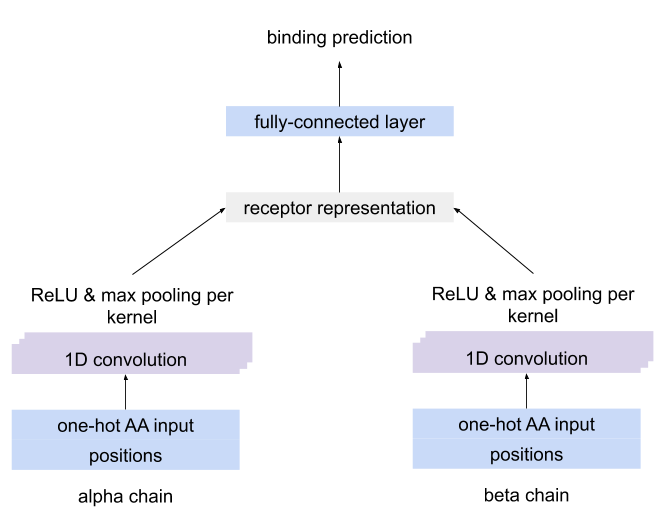
The architecture of the CNN for paired-chain receptor data
Requires one-hot encoded data as input (as produced by OneHot encoder).
Notes:
ReceptorCNN can only be used with ReceptorDatasets, it does not work with SequenceDatasets
ReceptorCNN can only be used for binary classification, not multi-class classification.
- Parameters
kernel_count (count) – number of kernels that will look for motifs for one chain
kernel_size (list) – sizes of the kernels = how many amino acids to consider at the same time in the chain sequence, can be a tuple of values; e.g. for value [3, 4] of kernel_size, kernel_count*len(kernel_size) kernels will be created, with kernel_count kernels of size 3 and kernel_count kernels of size 4 per chain
positional_channels (int) – how many positional channels where included in one-hot encoding of the receptor sequences (default is 3 in one-hot encoder)
sequence_type (SequenceType) – type of the sequence
device – which device to use for the model (cpu or gpu) - for more details see PyTorch documentation on device parameter
number_of_threads (int) – how many threads to use
random_seed (int) – number used as a seed for random initialization
learning_rate (float) – learning rate scaling the step size for optimization algorithm
iteration_count (int) – for how many iterations to train the model
l1_weight_decay (float) – weight decay l1 value for the CNN; encourages sparser representations
l2_weight_decay (float) – weight decay l2 value for the CNN; shrinks weight coefficients towards zero
batch_size (int) – how many receptors to process at once
training_percentage (float) – what percentage of data to use for training (the rest will be used for validation); values between 0 and 1
evaluate_at (int) – when to evaluate the model, e.g. every 100 iterations
background_probabilities – used for rescaling the kernel values to produce information gain matrix; represents the background probability of each amino acid (without positional information); if not specified, uniform background is assumed
YAML specification:
my_receptor_cnn: ReceptorCNN: kernel_count: 5 kernel_size: [3] positional_channels: 3 sequence_type: amino_acid device: cpu number_of_threads: 16 random_seed: 100 learning_rate: 0.01 iteration_count: 10000 l1_weight_decay: 0 l2_weight_decay: 0 batch_size: 5000
- can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- check_if_exists(path)[source]
The check_if_exists function checks if there is a stored model on the given path. Might be useful in the future for implementing checkpoints. See SklearnMethod for example usage.
- Parameters
path (Path) – path to folder where it should be checked if the model was stored previously
- Returns
whether the model was stored previously on the given path or not
- Return type
True/False
- fit(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label, cores_for_training: int = 2)[source]
The fit function fits the parameters of the machine learning model.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – if parallelization is available in the MLMethod (and the availability depends on the specific classifier), this is the number of processes that will be creating when fitting the model to speed up the computation.
- Returns
it doesn’t return anything, but fits the model parameters instead
- fit_by_cross_validation(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, number_of_splits: int = 5, label: Optional[immuneML.environment.Label.Label] = None, cores_for_training: int = - 1, optimization_metric=None)[source]
The fit_by_cross_validation function should implement finding the best model hyperparameters through cross-validation. In immuneML, preprocessing, encoding and ML hyperparameters can be optimized by using nested cross-validation (see TrainMLModelInstruction for more details). This function is in that setting the third level of nested cross-validation as it can optimize only over the model hyperparameters. It represents an alternative to optimizing the model hyperparameters in the TrainMLModelInstruction. Which one should be used depends on the use-case and specific models: models based on scikit-learn implementations come with this option by default (see SklearnMethod class), while custom classifiers typically do not implement this and just call fit() function and throw a warning instead.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
number_of_splits (int) – number of splits for the cross-validation to be performed for selection the best hyperparameters of the ML model; note that if this is used in combination with nested cross-validation in TrainMLModel instruction, it can result in very few examples in each split depending on the orginal dataset size and the nested cross-validation setup.
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – number of processes to be used during the cross-validation for model selection
optimization_metric (str) – the name of the optimization metric to be used to select the best model during cross-validation; when used with TrainMLModel instruction which is almost exclusively the case when the immuneML is run from the specification, this maps to the optimization metric in the instruction.
- Returns
it doesn’t return anything, but fits the model parameters instead
- get_class_mapping() dict[source]
Returns a dictionary containing the mapping between label values and values internally used in the classifier
- get_feature_names() list[source]
Returns the list of feature names (a list of strings) if available where the feature names were provided by the encoder in the EncodedData object.
- get_package_info() str[source]
Returns the package and version used for implementing the ML method if an external package was used or immuneML version if it is custom implementation. See py:mod:immuneML.ml_methods.SklearnMethod.SklearnMethod and py:mod:immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier for examples of both.
- get_params()[source]
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- load(path)[source]
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters
path (Path) – path to the folder where the model was stored using store() function
- Returns
it does not have a return value, but sets the attribute values of the object instead
- predict(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict function predicts the class for the given label across examples provided in encoded data.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label – (Label): the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
- Returns
e.g., {label_name: [class1, class2, class2, class1]}
- Return type
a dictionary where the key is the label_name and the value is a list of class predictions (one prediction per example)
- predict_proba(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict_proba function predicts class probabilities for the given label if the model supports probabilistic output. If not, it should raise a warning and return predicted classes without probabilities.
Note that when providing class probabilities the classes should have a specific (constant) order, and in case of binary classification, they should be ordered so that the negative class comes first and the positive one comes second. For this handling classes, see py:mod:immuneML.ml_methods.util.Util.Util.make_binary_class_mapping method that will automatically create class mapping for binary classification.
- Parameters
encoded_data (EncodedData) – an object of EncodedData class where the examples attribute should be used to make predictions. examples
format (attribute includes encoded examples in matrix) –
labels (provided here can include) –
labels) (been) –
function (so the labels attribute of the EncodedData object should NOT be used in this) –
set. (even if it is) –
label (Label) – the label for which the prediction should be made. It can be used to check if it matches the label that the
not (model has been trained for and if) –
any (an exception should be thrown. It is often an AssertionError as this can be checked before) –
made (prediction is) –
cases (but could also be a RuntimeError. It both) –
message. (it should include a user-friendly) –
- Returns
a dictionary where the key is the label name and the value a 2D numpy array with class probabilities of dimension [number_of_examples x number_of_classes_for_label], for instance for label CMV where the class can be either True or False and there are 3 examples to predict the class probabilities for: {CMV: [[0.2, 0.8], [0.55, 0.45], [0.98, 0.02]]}
- store(path: pathlib.Path, feature_names=None, details_path: Optional[pathlib.Path] = None)[source]
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters
path (Path) – path to folder where to store the model
feature_names (list) – list of feature names in the encoded data; this can be stored as well to make it easier to map linear models to specific features as provided by the encoded (e.g., in case of logistic regression, this feature list defines what coefficients refer to)
details_path (Path) – path to folder where to store the details of the model. The details can be there to better understand the model but are not mandatory and are typically not loaded with the model afterwards. This is user-friendly file that can be examined manually by the user. It does not have to be created or can be created at the same folder as the path parameters points to. In practice, when used with TrainMLModel instruction, this parameter will either be None or have the same value as path parameter.
- Returns
it does not have a return value
immuneML.ml_methods.SVC module
- class immuneML.ml_methods.SVC.SVC(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodThis is a wrapper of scikit-learn’s LinearSVC class. Please see the scikit-learn documentation of SVC for the parameters.
Note: if you are interested in plotting the coefficients of the SVC model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
my_svc: # user-defined method name SVC: # name of the ML method # sklearn parameters (same names as in original sklearn class) C: [0.01, 0.1, 1, 10, 100] # find the optimal value for C # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under SVC is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_svc: SVC
immuneML.ml_methods.SVM module
- class immuneML.ml_methods.SVM.SVM(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodThis is a wrapper of scikit-learn’s SVC class. Please see the scikit-learn documentation of SVC for the parameters.
Note: if you are interested in plotting the coefficients of the SVM model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
my_svm: # user-defined method name SVM: # name of the ML method # sklearn parameters (same names as in original sklearn class) C: [0.01, 0.1, 1, 10, 100] # find the optimal value for C kernel: linear # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under SVM is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_svm: SVM
immuneML.ml_methods.SklearnMethod module
- class immuneML.ml_methods.SklearnMethod.SklearnMethod(parameter_grid: Optional[dict] = None, parameters: Optional[dict] = None)[source]
Bases:
immuneML.ml_methods.MLMethod.MLMethodBase class for ML methods imported from scikit-learn. The classes inheriting SklearnMethod acting as wrappers around imported ML methods from scikit-learn have to implement:
the __init__() method,
get_params(label) and
_get_ml_model()
Other methods can also be overwritten if needed. The arguments and specification described bellow applied for all classes inheriting SklearnMethod.
- Parameters
parameters – a dictionary of parameters that will be directly passed to scikit-learn’s class upon calling __init__() method; for detailed list see scikit-learn’s documentation of the specific class inheriting SklearnMethod
parameter_grid – a dictionary of parameters which all have to be valid arguments for scikit-learn’s corresponding class’ __init__() method (same as parameters), but unlike parameters argument can contain list of values instead of one value; if this is specified and “model_selection_cv” is True (in the specification) or just if fit_by_cross_validation() is called, a grid search will be performed over these parameters and the optimal model will be kept
YAML specification:
- ml_methods:
- log_reg:
- LogisticRegression: # name of the class inheriting SklearnMethod
# sklearn parameters (same names as in original sklearn class) max_iter: 1000 # specific parameter value penalty: l1 # Additional parameter that determines whether to print convergence warnings show_warnings: True
# if any of the parameters under LogisticRegression is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5
- svm_with_cv:
- SVM: # name of another class inheriting SklearnMethod
# sklearn parameters (same names as in original sklearn class) alpha: 10 # Additional parameter that determines whether to print convergence warnings show_warnings: True
# no grid search will be done model_selection_cv: False
- FIT = 'fit'
- FIT_CV = 'fit_CV'
- can_predict_proba() bool[source]
Returns whether the ML model can be used to predict class probabilities or class assignment only.
- check_if_exists(path: pathlib.Path)[source]
The check_if_exists function checks if there is a stored model on the given path. Might be useful in the future for implementing checkpoints. See SklearnMethod for example usage.
- Parameters
path (Path) – path to folder where it should be checked if the model was stored previously
- Returns
whether the model was stored previously on the given path or not
- Return type
True/False
- fit(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label, cores_for_training: int = 2)[source]
The fit function fits the parameters of the machine learning model.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – if parallelization is available in the MLMethod (and the availability depends on the specific classifier), this is the number of processes that will be creating when fitting the model to speed up the computation.
- Returns
it doesn’t return anything, but fits the model parameters instead
- fit_by_cross_validation(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, number_of_splits: int = 5, label: Optional[immuneML.environment.Label.Label] = None, cores_for_training: int = - 1, optimization_metric='balanced_accuracy')[source]
The fit_by_cross_validation function should implement finding the best model hyperparameters through cross-validation. In immuneML, preprocessing, encoding and ML hyperparameters can be optimized by using nested cross-validation (see TrainMLModelInstruction for more details). This function is in that setting the third level of nested cross-validation as it can optimize only over the model hyperparameters. It represents an alternative to optimizing the model hyperparameters in the TrainMLModelInstruction. Which one should be used depends on the use-case and specific models: models based on scikit-learn implementations come with this option by default (see SklearnMethod class), while custom classifiers typically do not implement this and just call fit() function and throw a warning instead.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
number_of_splits (int) – number of splits for the cross-validation to be performed for selection the best hyperparameters of the ML model; note that if this is used in combination with nested cross-validation in TrainMLModel instruction, it can result in very few examples in each split depending on the orginal dataset size and the nested cross-validation setup.
label (Label) – the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
cores_for_training (int) – number of processes to be used during the cross-validation for model selection
optimization_metric (str) – the name of the optimization metric to be used to select the best model during cross-validation; when used with TrainMLModel instruction which is almost exclusively the case when the immuneML is run from the specification, this maps to the optimization metric in the instruction.
- Returns
it doesn’t return anything, but fits the model parameters instead
- get_class_mapping() dict[source]
Returns a dictionary containing the mapping between label values and values internally used in the classifier
- get_feature_names() list[source]
Returns the list of feature names (a list of strings) if available where the feature names were provided by the encoder in the EncodedData object.
- get_package_info() str[source]
Returns the package and version used for implementing the ML method if an external package was used or immuneML version if it is custom implementation. See py:mod:immuneML.ml_methods.SklearnMethod.SklearnMethod and py:mod:immuneML.ml_methods.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier for examples of both.
- abstract get_params()[source]
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- load(path: pathlib.Path, details_path: Optional[pathlib.Path] = None)[source]
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters
path (Path) – path to the folder where the model was stored using store() function
- Returns
it does not have a return value, but sets the attribute values of the object instead
- predict(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict function predicts the class for the given label across examples provided in encoded data.
- Parameters
encoded_data (EncodedData) – an instance of EncodedData class which includes encoded examples (repertoires, receptors or sequences), their labels, names of the features and other additional information. Most often, only examples and labels will be used. Examples are either a dense numpy matrix or a sparse matrix, where columns correspond to features and rows correspond to examples. There are a few encodings which make multidimensional outputs that do not follow this pattern, but they are tailored to specific ML methods which require such input (for instance, one hot encoding and ReceptorCNN method).
label – (Label): the label for which the classifier will be created. immuneML also supports multi-label classification, but it is handled outside MLMethod class by creating an MLMethod instance for each label. This means that each MLMethod should handle only one label.
- Returns
e.g., {label_name: [class1, class2, class2, class1]}
- Return type
a dictionary where the key is the label_name and the value is a list of class predictions (one prediction per example)
- predict_proba(encoded_data: immuneML.data_model.encoded_data.EncodedData.EncodedData, label: immuneML.environment.Label.Label)[source]
The predict_proba function predicts class probabilities for the given label if the model supports probabilistic output. If not, it should raise a warning and return predicted classes without probabilities.
Note that when providing class probabilities the classes should have a specific (constant) order, and in case of binary classification, they should be ordered so that the negative class comes first and the positive one comes second. For this handling classes, see py:mod:immuneML.ml_methods.util.Util.Util.make_binary_class_mapping method that will automatically create class mapping for binary classification.
- Parameters
encoded_data (EncodedData) – an object of EncodedData class where the examples attribute should be used to make predictions. examples
format (attribute includes encoded examples in matrix) –
labels (provided here can include) –
labels) (been) –
function (so the labels attribute of the EncodedData object should NOT be used in this) –
set. (even if it is) –
label (Label) – the label for which the prediction should be made. It can be used to check if it matches the label that the
not (model has been trained for and if) –
any (an exception should be thrown. It is often an AssertionError as this can be checked before) –
made (prediction is) –
cases (but could also be a RuntimeError. It both) –
message. (it should include a user-friendly) –
- Returns
a dictionary where the key is the label name and the value a 2D numpy array with class probabilities of dimension [number_of_examples x number_of_classes_for_label], for instance for label CMV where the class can be either True or False and there are 3 examples to predict the class probabilities for: {CMV: [[0.2, 0.8], [0.55, 0.45], [0.98, 0.02]]}
- store(path: pathlib.Path, feature_names=None, details_path: Optional[pathlib.Path] = None)[source]
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters
path (Path) – path to folder where to store the model
feature_names (list) – list of feature names in the encoded data; this can be stored as well to make it easier to map linear models to specific features as provided by the encoded (e.g., in case of logistic regression, this feature list defines what coefficients refer to)
details_path (Path) – path to folder where to store the details of the model. The details can be there to better understand the model but are not mandatory and are typically not loaded with the model afterwards. This is user-friendly file that can be examined manually by the user. It does not have to be created or can be created at the same folder as the path parameters points to. In practice, when used with TrainMLModel instruction, this parameter will either be None or have the same value as path parameter.
- Returns
it does not have a return value
immuneML.ml_methods.TCRdistClassifier module
- class immuneML.ml_methods.TCRdistClassifier.TCRdistClassifier(percentage: float, show_warnings: bool = True)[source]
Bases:
immuneML.ml_methods.SklearnMethod.SklearnMethodImplementation of a nearest neighbors classifier based on TCR distances as presented in Dash P, Fiore-Gartland AJ, Hertz T, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017; 547(7661):89-93. doi:10.1038/nature22383.
This method is implemented using scikit-learn’s KNeighborsClassifier with k determined at runtime from the training dataset size and weights linearly scaled to decrease with the distance of examples.
- Parameters
percentage (float) – percentage of nearest neighbors to consider when determining receptor specificity based on known receptors (between 0 and 1)
show_warnings (bool) – whether to show warnings generated by scikit-learn, by default this is True.
YAML specification:
my_tcr_method: TCRdistClassifier: percentage: 0.1 show_warnings: True