Quickstart: Galaxy with button-based tools
This tutorial will show you how to use immuneML for a simple machine learning analysis on an adaptive immune receptor repertoire (AIRR) dataset. This example dataset consists of 100 synthetic immune repertoires (amino acid sequences generated by OLGA), each containing 1000 CDR3 sequences. In half the repertoires, the subsequence ‘VLEQ’ has been implanted in 5% of the CDR3 sequences to simulate a disease signal. Using immuneML, we will encode the data as 3-mer frequencies and train a logistic regression model to predict the disease status of each repertoire (i.e., whether the repertoire contains the disease signal ‘VLEQ’), which is a binary classification problem.
Getting started through Galaxy
The Galaxy web interface is available at https://galaxy.immuneml.uio.no/. You may choose to register a user account or perform the analysis as an anonymous user.
Different functionalities are available as Galaxy tools (left menu), and the analysis results appear in the Galaxy history (right list). These history elements can be used as input for the next tool, creating a multi-step analysis workflow.
The purpose of the Quickstart tutorial is to quickly guide you through an analysis step by step, without assuming prior knowledge of Galaxy. Additionally, you can check out our Introduction to Galaxy for a more detailed explanation of the Galaxy interface.
Step 1: importing the dataset to a Galaxy history
Every immuneML analysis takes a dataset as input. For the Quickstart tutorial, an example dataset has been prepared and is available through this Galaxy history (under ‘Shared data’ > ‘Histories’ > ‘Quickstart data’). Alternatively, the tutorial How to make an immuneML dataset in Galaxy describes in detail how to make an immuneML dataset using your own data.
This Quickstart dataset Galaxy history contains the following items:
100 repertoire .tsv files in AIRR format. For details about the AIRR format, see the AIRR documentation and this example file).
A Collection of repertoires. This history element collects the 100 above-mentioned repertoire files in a Galaxy collection. This Galaxy collection makes it easier to select the repertoires as an input to Galaxy tools (instead of selecting all 100 files manually, you can select the collection). To read how to make your own Galaxy collection, see Creating a Galaxy collection of files.
A metadata.csv file. The metadata file describes which of the 100 repertoires are diseased and healthy, under the column named ‘signal_disease’ which contains the values True and False. For details about the metadata file, see What should the metadata file look like? and this example file
Individual files can be inspected by clicking the eyeball icons. To import the complete history, click the + icon in the right upper corner.
Step 2: creating an immuneML Galaxy dataset
Use the Create dataset Galaxy tool (under ‘immuneML tools’) to import the dataset and create an immuneML dataset history item, which can subsequently be used as input for other Galaxy tools.
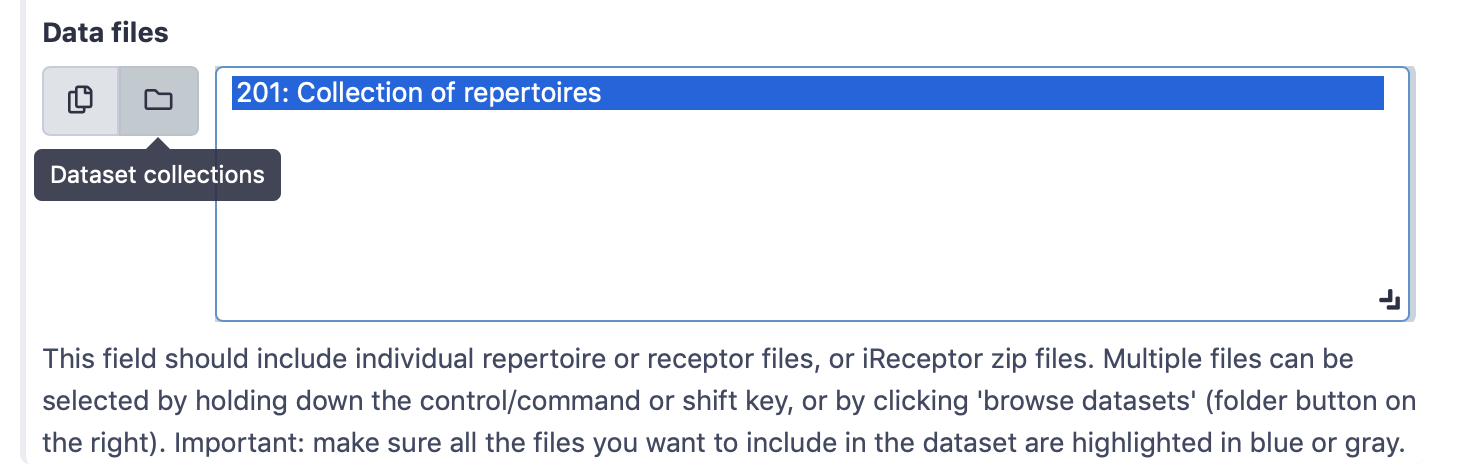
Select ‘Simplified interface’, then ‘repertoire dataset’ as dataset type and ‘AIRR’ data format, and select the metadata.csv file as metadata file. Under the ‘Data files’ field, all repertoire files must be selected. This can either be done by manually selecting all 100 repertoire files, or by selecting the Collection of repertoires. For convenience, we will use the latter option.
By default, the ‘Data files’ menu will show all txt-like files (such as repertoire files) that are present in the history. By clicking the ‘Dataset collections’ button (folder icon), the menu will instead only show the collections in the history. Select the collection of repertoires so it becomes highlighted.
Finally, click ‘execute’.
Three new items will appear in the Galaxy history. If everything went correctly, the items would appear green. If the items are red, an error occurred (check if you correctly selected the collection of repertoires).
By viewing ‘Summary: dataset generation’ (click the eyeball icon) you can find details about the newly generated dataset, including the name of the dataset, the dataset type and size, and a download link.
The next item, ‘create_dataset.yaml’ the YAML specification file that was used by immuneML to create the dataset. This YAML specification could be used when running the Create dataset tool with the ‘Advanced’ interface.
Finally, ‘immuneML dataset’ is a new Galaxy collection containing the immuneML dataset in the ImmuneML format. The ImmuneML format is not human-readable, but it ensures that you can quickly import the dataset into various Galaxy tools without having to repeatedly specify the import parameters.
Step 3: running the analysis
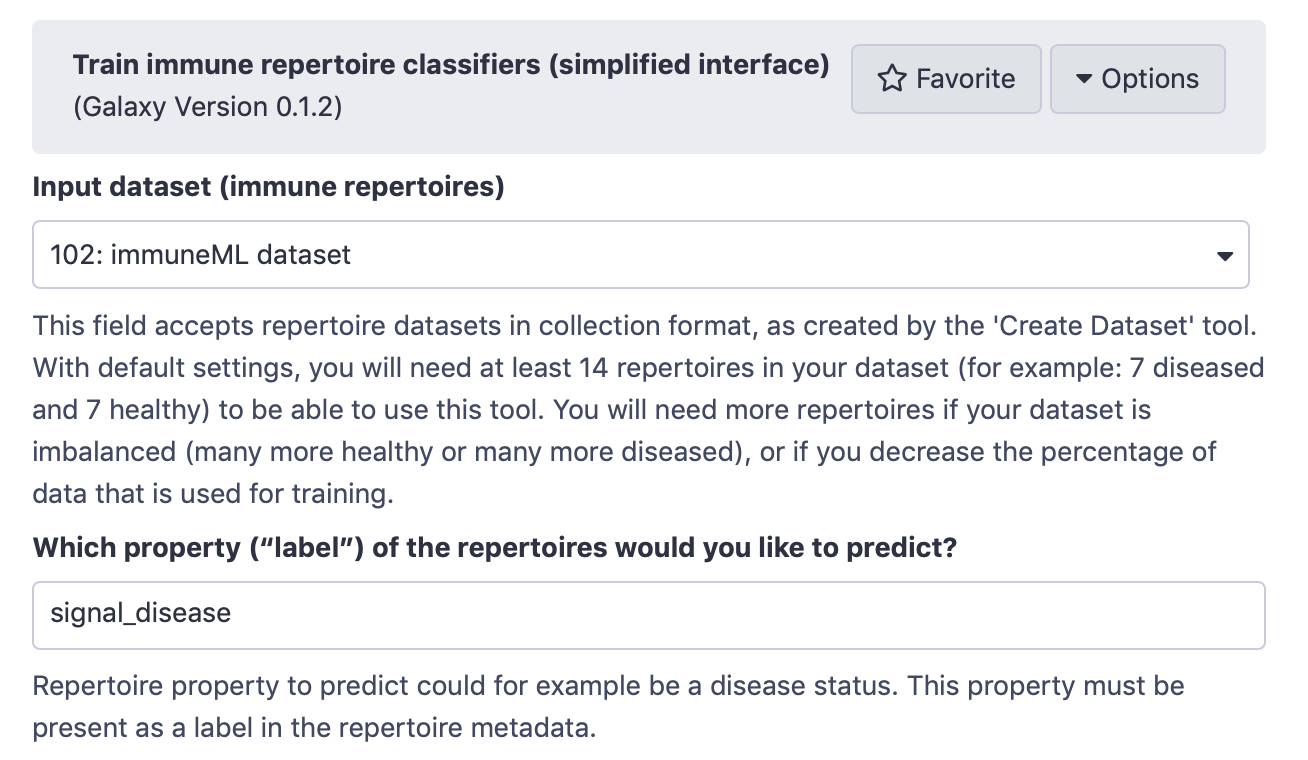
The immuneML Galaxy instance has two tools that provide a simplified interface for training immune receptor and immune repertoire classifiers. Here, we will use the Galaxy tool Train immune repertoire classifiers (simplified interface) (under ‘immuneML tools’).
Select the immuneML dataset as input dataset, and write the name of the label that you want to predict. In this case, the label is ‘signal_disease’, because it is defined in the metadata.csv file.
All other options are configurable however you want (choose at least one ML method). To speed up the running time, decrease the number of times to repeat the training process with random splits of data (last question), for example by setting the value to 1. When using 1 data split, selecting all ML methods and otherwise default settings, the running time is expected to be approximately 2 minutes.
For background information explaining the relevant machine learning concepts, please refer to the text written below the tool (the text can also be found in the documentation).
Step 4: understanding the results
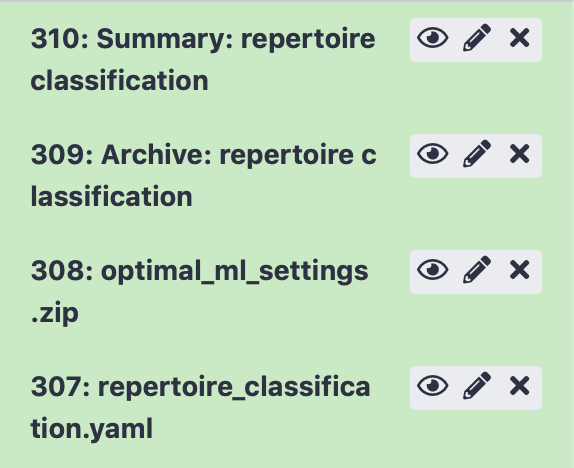
After running the Galaxy tool for training ML models, four new items will appear in the Galaxy history.
The history item ‘Summary: repertoire classification’ contains HTML pages displaying a summary of the analysis. On the first page, you will find a table which shows for each of the splits (i.e., each repetition of the training process) which of the machine learning models performed best during cross-validation. Furthermore, there is a barplot comparing the performance of the different machine learning models on the test set across all the splits.
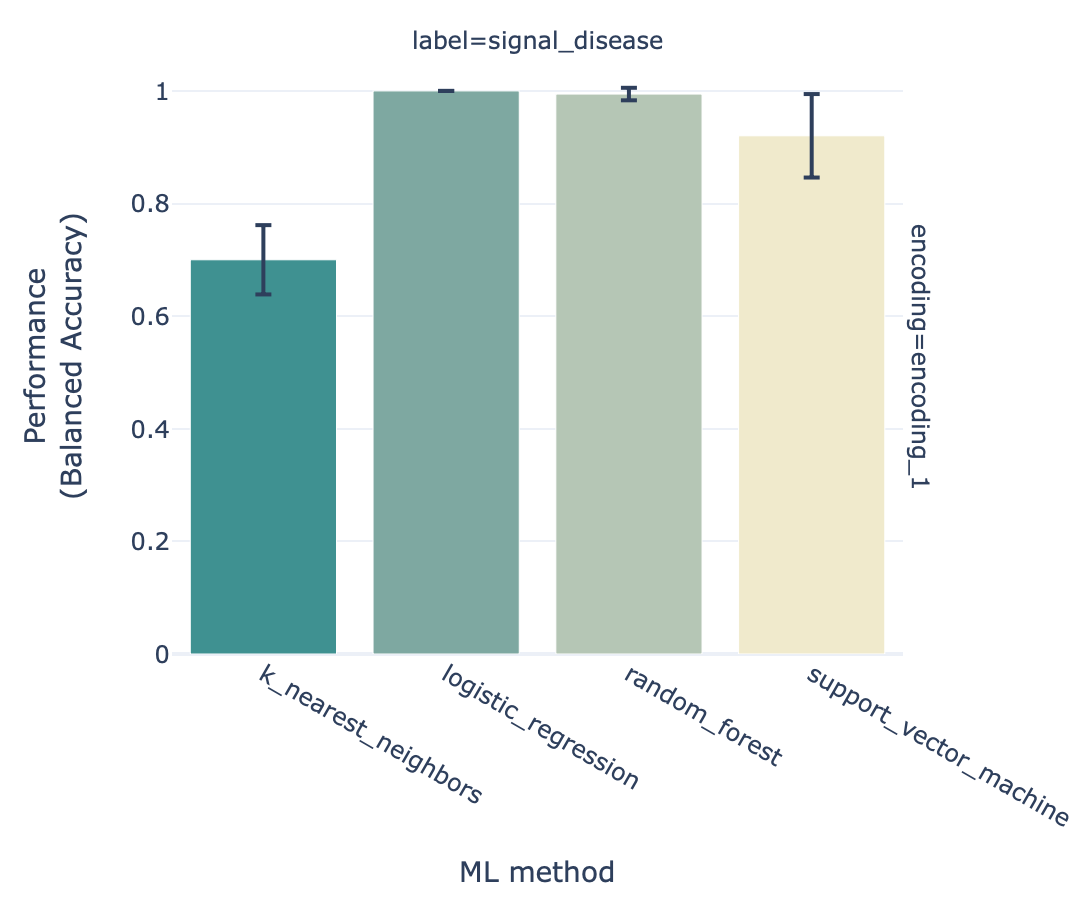
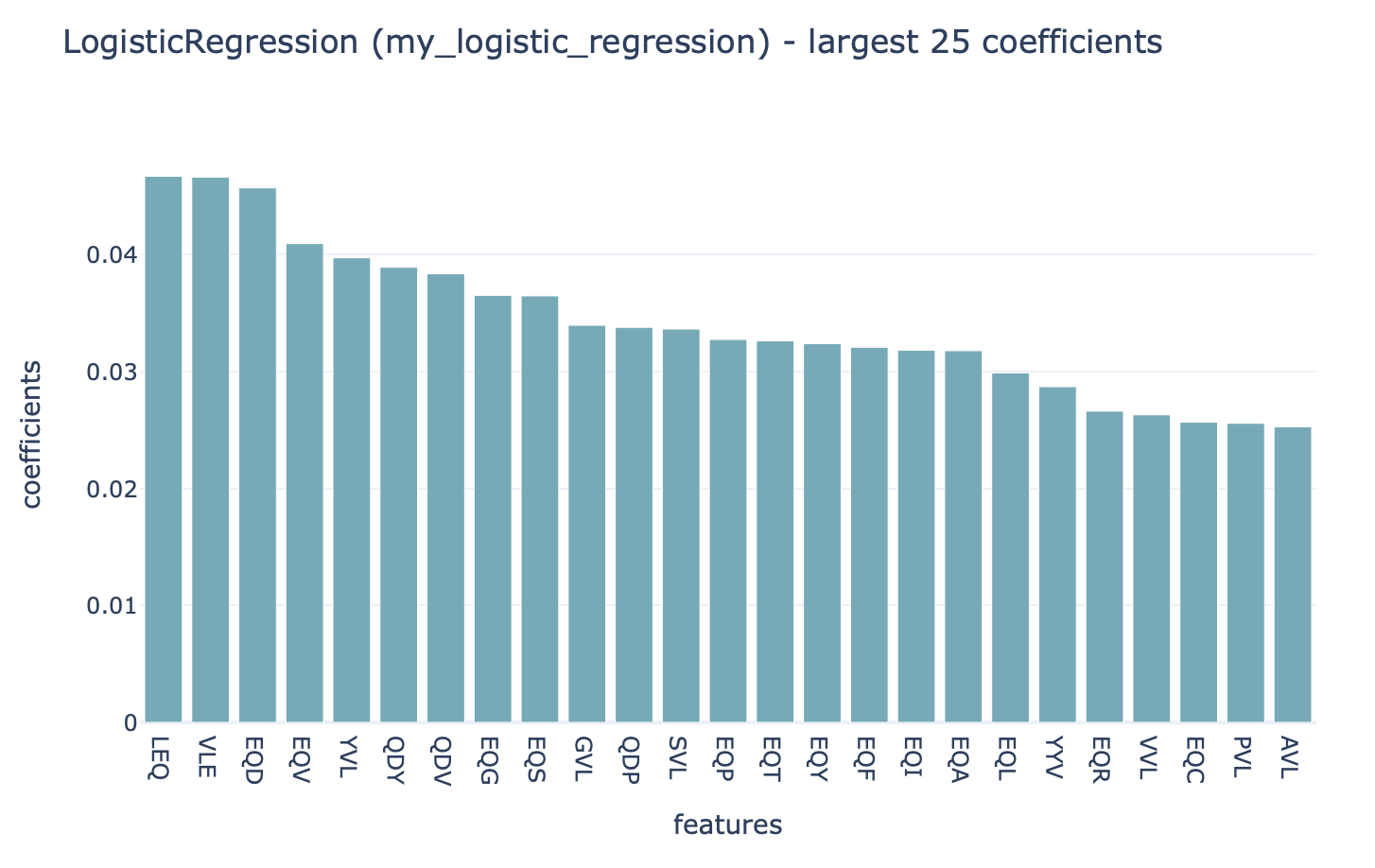
You can click ‘see details’ in the table to find a more detailed explanation about how well each model performed in the inner loop of cross validation (on the validation sets). By clicking ‘see reports’ you can find reports showing the 25 largest coefficients of logistic regression or support vector machine, or the top 25 most important features for random forest. Below is an example of logistic regression coefficients. Notice how the coefficients with the highest values are associated with the k-mers ‘VLE’ and ‘LEQ’, which overlap with the implanted disease signal ‘VLEQ’, meaning the ML model learned the correct signal.
The next item, ‘Archive: repertoire classification’ contains a downloadable archive of the complete immuneML ouput (including the files available through the other history elements).
The history element ‘optimal_ml_settings.zip’ is a .zip file containing the configuration of the optimal ML settings, including settings for the encoding and machine learning method. Using the YAML-based Galaxy tool Apply machine learning models to new data the trained ML model can be used to make predictions on a new dataset where the true disease labels are not known.
Finally, ‘repertoire_classification.yaml’ contains the YAML specification that was used to run the immuneML analysis. This YAML specification was automatically generated based on the options you selected in the interface. You may also use this YAML specification file as a starting point for defining your own analyses later on.
What’s next?
If you haven’t done it already, it is highly recommended to follow the Introduction to Galaxy. If you want to try running immuneML on your own dataset, be sure to check out How to make an immuneML dataset in Galaxy.
While in this tutorial we trained an ML model for making a prediction per repertoire, immuneML also provides a button-based interface for training ML models to make predictions per sequence, such as antigen binding: Train immune receptor classifiers (simplified interface).
immuneML provides many more options for customizing your machine learning analysis when using the YAML-based specification. To get started with this, you can try out Quickstart: Galaxy with YAML-based tools, for example using the YAML specification that was produced in the Galaxy history (‘repertoire_classification.yaml’) with some minor modifications. See also the tutorial How to specify an analysis with YAML to understand how the YAML specification can be altered.
Other tutorials for how to use each of the (YAML-based) immuneML Galaxy tools can be found under immuneML & Galaxy.
The Galaxy interface is intended to make it easy for users to try out immuneML quickly, but for large-scale analyses, please install immuneML locally or on a private server.