immuneML execution flow¶
immuneML is run by providing the analysis specification and the output path. This section describes what happens internally when immuneML is run.
Note: this is advanced developer documentation, these details are not crucial to understand for adding new components (MLMethod, Encoder, Report classes) to immuneML.
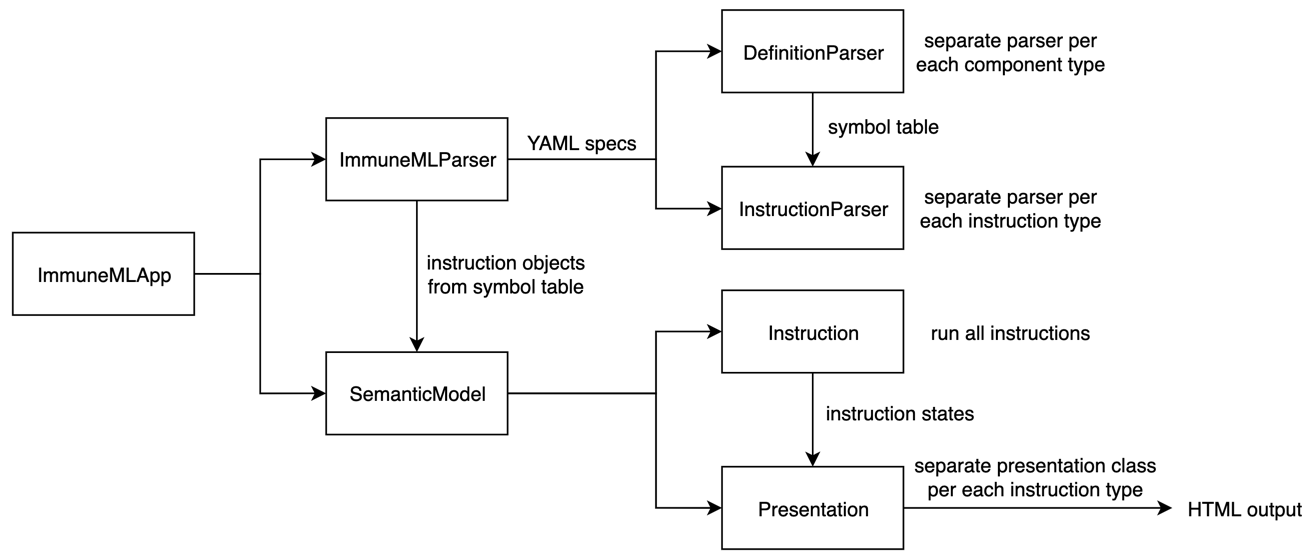
Overview of classes involved in running immuneML¶
When immuneML is run from the command line, the immune-ml command is mapped to the main() method in ImmuneMLApp.py. The ImmuneMLApp class is then instantiated with the specification path and output path. Running immuneML corresponds to parsing the specification file, running the specified analyses and constructing the HTML output.
Parsing the specification¶
To parse the specification file, ImmuneMLApp calls ImmuneMLParser from the immuneML.dsl
(domain-specific language) package. All parsing functionality is
located in that package. Parsing consists of two major steps again: parsing the definitions component (motifs, signals, encodings, datasets,
ML methods, reports, etc) and parsing the instructions (concrete analyses to be performed using the parsed components). This maps to two classes:
DefinitionParser and InstructionParser.
DefinitionParser parses each of the components and invokes a separate parser class (e.g., EncodingParser, MLParser) for each component.
Parsing entails reading the YAML, and determining which analysis components are specified with which parameters.
The result is a set of objects and classes which are stored in a SymbolTable.
The SymbolTable functions like a dictionary where all analysis components are stored.
For instance, if a user has specified a report like so:
definitions:
reports:
my_report: SequenceLengthDistribution:
... # this is a snippet of a larger yaml file
Then the SymbolTable will contain a SequenceLengthDistribution object which can be retrieved through the
name (symbol) ‘my_report’.
When all components are parsed, the filled symbol table is passed as input to the InstructionParser class.
It uses the objects from the symbol table to populate the Instruction objects that will run the analysis.
Again, each instruction type has its own parser (e.g., TrainMLModelParser, ExploratoryAnalysisParser).
The specific parsers were created to ensure user-friendly error messages in case of misspecified YAML files.
Creating the semantic model and running instructions¶
Once the YAML specification is parsed, all instruction objects are extracted and forwarded to the semantic model. The object of the SemanticModel
class will then run all instructions one by one, collect their states as the output of each instruction and use them to generate the presentation for
the user. Each instruction class is a subclass of Instruction class and implements run() method and returns the instruction’s state object as output.
The instruction state objects consist of input data and parameters for the instruction and the instruction results (added to the state during the execution).
This is then used as input for the presentation part where the results are shown to the user. The code for the presentation
part is located in the immuneML.presentation package and are grouped by presentation format. The only format supported at the moment is HTML.
Constructing the HTML output¶
To generate the HTML output from the analyses, the semantic model finds the presentation class that corresponds to the instruction and calls its build() method with the instructions state as input. This method creates a mapping between the state object and the information that will be shown in the HTML files. For each instruction, a set of presentation templates is available. The templates are filled from the generated mapping using the pystache package. This process is repeated for each instruction if there are multiple ones and a common index.html file is generated linking to all HTML files. If there is only one instruction, the index.html file is the mapped HTML template of that instruction.