Information for new developers¶
Besides the challenge of developing a new ML method, setting up the computational pipelines to thoroughly (and correctly) benchmark the new method against existing solutions is a large and complex task. And in practice, independently developed methods may be evaluated under different conditions, resulting in a lack of transparency regarding which method(s) are optimal for a given prediction problem.
We therefore designed immuneML as a platform to support and accelerate the development of AIRR-ML methods and benchmarking thereof. Integration into immuneML has benefits both for new and existing AIRR-ML methods.
We highly encourage anyone to integrate their method into immuneML using the provided tutorials. If you experience any issues or have additional questions, please feel free to reach out to contact@immuneml.uio.no and we will be happy to help!
Why should I integrate my method into immuneML?¶
If you are designing a new AIRR-ML method from scratch, developing the method directly as a module inside immuneML can save you a lot of time and boilerplate code. And even for already existing methods, integrating it into immuneML directly allows efficient and transparent benchmarking against other methods, and makes your tool just as easily available for other AIRR-ML researchers interested in benchmarking.
The following tasks have already been implemented (and thoroughly tested) in immuneML and can directly be reused:
Importing of datasets in a large variety of formats
Data preprocessing
Splitting data into training, validation and test sets (nested cross-validation)
Encoding data into a variety of different formats, including one-hot or k-mer based encodings
Training and hyperparameter optimisation of ML models
Performance evaluation of models through reports, which may be used directly as results figures
A range of different baseline or ‘competitor’ algorithms to benchmark your new model against
Fast benchmarking of ML models through smart internal data representations, parallelization and caching
Exact result reproducibility through a YAML specification file containing all model and benchmarking parameters
Which only leaves the following to be implemented:
A new
MLMethodclassA new
DatasetEncoderclass, if none of the existing encodings sufficeOptionally, one or more
Reportclasses for plotting model-specific results
How do I get started integrating my method into immuneML?¶
High-level algorithm design¶
Within immuneML, ML settings consisting of data preprocessing (optional), encoding and ML method (along with a concrete set of hyperparameters) are being evaluated and compared.
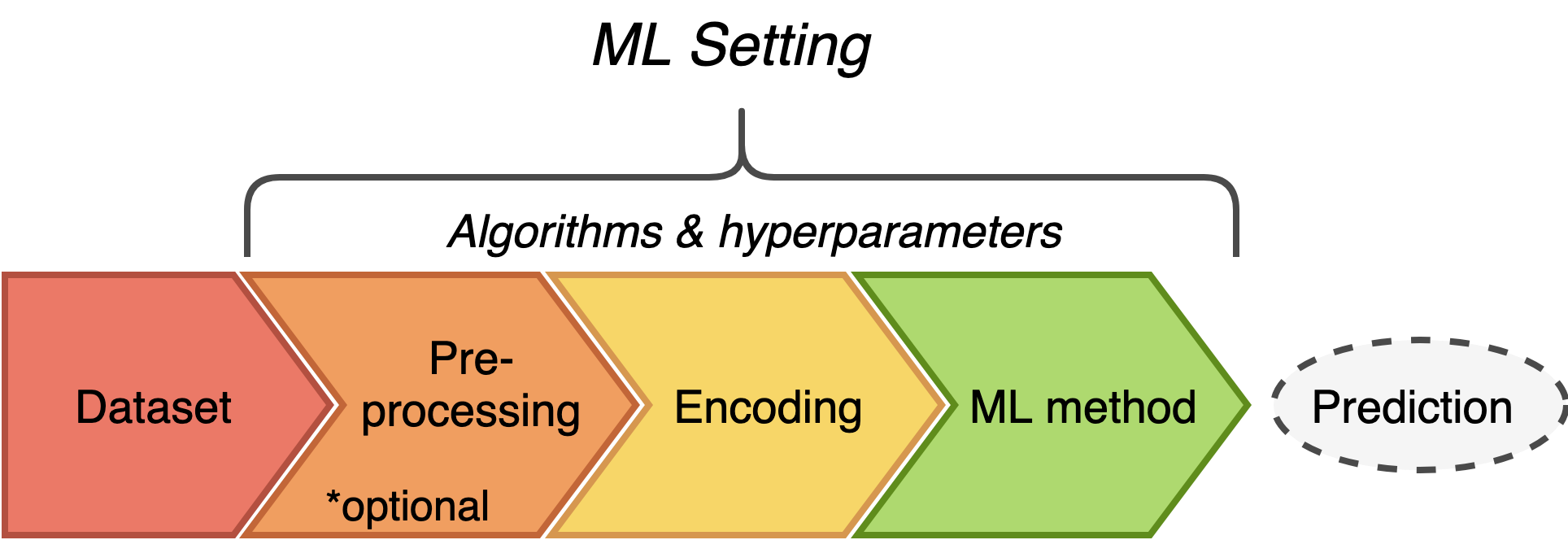
Thus, the first step of integrating your method into immuneML is to decide which parts of your algorithm belong to each step. Since preprocessing steps are usually trivial to separate and not commonly used, the next section will focus on the difference between encoding and ML method steps.
Designing an encoding¶
An encoding (sometimes called ‘design matrix’ or input matrix ‘X’) is a numeric representation of the data. We highly recommend you take a look at the existing Encodings to see if an appropriate encoding already exists. For example, deep learning methods for AIRR data often require sequences to be represented by OneHot encoding. Furthermore, k-mer based encodings (KmerFrequency), or distance matrices (Distance, CompAIRRDistance) are commonly used.
For other methods, the separation between encoding and ML method is less trivial. Perhaps some specific, complex algorithmic steps need to be performed before arriving at the final encoded data. In those cases, it may be necessary to design your own encoding. Depending on your specific method, there may be multiple valid ways of dividing the algorithm into encoding and ML method, but the encoded data format should adhere the following:
The encoded data is a multidimensional matrix
This matrix contains exclusively numeric values
The first dimension represents the examples ( = individual repertoires, receptors or sequences). In other words, the encoding gives some numeric representation per example.
There are usually two dimensions (examples x features), but multiple additional feature dimensions may be present (examples x features1 x features2 x features3 …)
Designing an ML method¶
An ML method takes in an encoded data matrix, fits a model and is able to make predictions on new data.
The dataset contains labels with two or more classes.
For example, a sequence dataset can have label “celiac” and classes “sick” and “healthy”.
ML models added to immuneML may be binary classifiers (2 classes) or support more classes,
but a model is always trained for only one label at a time. If multiple labels are specified
during training (e.g., sick/healthy for multiple different diseases), the model is trained for only one label at a time.
It is currently not yet possible to use immuneML for regression (predicting numeric values instead of classes).
Next steps¶
Follow the tutorial Set up immuneML for development
Familiarise yourself with the immuneML data model. Decide whether your encoding and/or ML method should be implemented for one or multiple data types.
If a new encoder is needed, follow the tutorial How to add a new encoding.
If a new ML method is needed, follow the tutorial How to add a new machine learning method
If your method or report requires some specific reports, follow the tutorial How to add a new report