immuneML.ml_methods.classifiers package¶
Submodules¶
immuneML.ml_methods.classifiers.AtchleyKmerMILClassifier module¶
immuneML.ml_methods.classifiers.BinaryFeatureClassifier module¶
- class immuneML.ml_methods.classifiers.BinaryFeatureClassifier.BinaryFeatureClassifier(training_percentage: float = None, random_seed: int = None, max_features: int = None, patience: int = None, min_delta: float = None, keep_all: bool = None, result_path: Path = None)[source]¶
Bases:
MLMethodA simple classifier that takes in encoded data containing features with only 1/0 or True/False values.
This classifier gives a positive prediction if any of the binary features for an example are ‘true’. Optionally, the classifier can select an optimal subset of these features. In this case, the given data is split into a training and validation set, a minimal set of features is learned through greedy forward selection, and the validation set is used to determine when to stop growing the set of features (earlystopping). Earlystopping is reached when the optimization metric on the validation set no longer improves for a given number of features (patience). The optimization metric is the same metric as the one used for optimization in the
TrainMLModelInstruction.Currently, this classifier can be used in combination with two encoders:
The classifier can be used in combination with the
MotifEncoder, such that sequences containing any of the positive class-associated motifs are classified as positive. A reduced subset of binding-associated motifs can be learned (when keep_all is false). This results in a set of complementary motifs, minimizing the redundant predictions made by different motifs.Alternatively, this classifier can be combined with the
SimilarToPositiveSequenceEncodersuch that any sequence that falls within a given hamming distance from any of the positive class sequences in the training set are classified as positive. Parameter keep_all should be set to true, since this encoder creates only 1 feature.
Specification arguments:
training_percentage (float): What percentage of data to use for training (the rest will be used for validation); values between 0 and 1
keep_all (bool): Whether to keep all the input features (true) or learn a reduced subset (false). By default, keep_all is false.
random_seed (int): Random seed for splitting the data into training and validation sets when learning a minimal subset of features. This is only used when keep_all is false.
max_features (int): The maximum number of features to allow in the reduced subset. When this number is reached, no more features are added even if the earlystopping criterion is not reached yet. This is only used when keep_all is false. By default, max_features is 100.
patience (int): The patience for earlystopping. When earlystopping is reached, <patience> more features are added to the reduced set to test whether the optimization metric on the validation set improves again. By default, patience is 5.
min_delta (float): The delta value used to test if there was improvement between the previous set of features and the new set of features (+1). By default, min_delta is 0, meaning the new set of features does not need to yield a higher optimization metric score on the validation set, but it needs to be at least equally high as the previous set.
YAML specification:
definitions: ml_methods: my_motif_classifier: MotifClassifier: training_percentage: 0.7 max_features: 100 patience: 5 min_delta: 0 keep_all: false
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_params(for_refitting=False)[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- load(path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.DeepRC module¶
- class immuneML.ml_methods.classifiers.DeepRC.DeepRC(validation_part, add_positional_information, kernel_size, n_kernels, n_additional_convs, n_attention_network_layers, n_attention_network_units, n_output_network_units, consider_seq_counts, sequence_reduction_fraction, reduction_mb_size, n_updates, n_torch_threads, learning_rate, l1_weight_decay, l2_weight_decay, evaluate_at, sample_n_sequences, training_batch_size, n_workers, sequence_counts_scaling_fn, keep_dataset_in_ram, pytorch_device_name)[source]¶
Bases:
MLMethodThis classifier uses the DeepRC method for repertoire classification. The DeepRC ML method should be used in combination with the DeepRC encoder. Also consider using the DeepRCMotifDiscovery report for interpretability.
Notes:
DeepRC uses PyTorch functionalities that depend on GPU. Therefore, DeepRC does not work on a CPU.
This wrapper around DeepRC currently only supports binary classification.
Reference: Michael Widrich, Bernhard Schäfl, Milena Pavlović, Geir Kjetil Sandve, Sepp Hochreiter, Victor Greiff, Günter Klambauer ‘DeepRC: Immune repertoire classification with attention-based deep massive multiple instance learning’. bioRxiv preprint doi: https://doi.org/10.1101/2020.04.12.038158
Specification arguments:
validation_part (float): the part of the data that will be used for validation, the rest will be used for training.
add_positional_information (bool): whether positional information should be included in the input features.
kernel_size (int): the size of the 1D-CNN kernels.
n_kernels (int): the number of 1D-CNN kernels in each layer.
n_additional_convs (int): Number of additional 1D-CNN layers after first layer
n_attention_network_layers (int): Number of attention layers to compute keys
n_attention_network_units (int): Number of units in each attention layer
n_output_network_units (int): Number of units in the output layer
consider_seq_counts (bool): whether the input data should be scaled by the receptor sequence counts.
sequence_reduction_fraction (float): Fraction of number of sequences to which to reduce the number of sequences per bag based on attention weights. Has to be in range [0,1].
reduction_mb_size (int): Reduction of sequences per bag is performed using minibatches of reduction_mb_size` sequences to compute the attention weights.
n_updates (int): Number of updates to train for
n_torch_threads (int): Number of parallel threads to allow PyTorch
learning_rate (float): Learning rate for adam optimizer
l1_weight_decay (float): l1 weight decay factor. l1 weight penalty will be added to loss, scaled by l1_weight_decay
l2_weight_decay (float): l2 weight decay factor. l2 weight penalty will be added to loss, scaled by l2_weight_decay
sequence_counts_scaling_fn: it can either be log (logarithmic scaling of sequence counts) or None
sequence_counts_scaling_fn: it can either be log (logarithmic scaling of sequence counts) or None
evaluate_at (int): Evaluate model on training and validation set every evaluate_at updates. This will also check for a new best model for early stopping.
sample_n_sequences (int): Optional random sub-sampling of sample_n_sequences sequences per repertoire. Number of sequences per repertoire might be smaller than sample_n_sequences if repertoire is smaller or random indices have been drawn multiple times. If None, all sequences will be loaded for each repertoire.
training_batch_size (int): Number of repertoires per minibatch during training.
n_workers (int): Number of background processes to use for converting dataset to hdf5 container and training set data loader.
pytorch_device_name (str): The name of the pytorch device to use. This name will be passed to torch.device(self.pytorch_device_name). The default value is cuda:0
YAML specification:
definitions: ml_methods: my_deeprc_method: DeepRC: validation_part: 0.2 add_positional_information: True kernel_size: 9
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- fit_by_cross_validation(encoded_data: EncodedData, number_of_splits: int = 5, label: Label = None, cores_for_training: int = -1, optimization_metric=None)[source]¶
See also: _fit_by_cross_validation.
This method should not be overwritten.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_package_info() str[source]¶
Should return the relevant version numbers of immuneML and all external packages that were used for the immuneML implementation. This information will be exported and is required for transparency and reproducibility.
This method should be overwritten to add any additional packages if necessary. For instance, versions of scikit-learn.
- get_params(for_refitting=False)[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- load(path: Path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- make_data_loader(full_dataset, indices, label_name, eval_only: bool, is_train: bool, n_workers=1)[source]¶
Creates a pytorch dataloader using DeepRC’s RepertoireDataReaderBinary
- Parameters:
hdf5_filepath – the path to the HDF5 file
pre_loaded_hdf5_file – Optional: It is faster to load the hdf5 file into the RAM as dictionary instead of keeping it on the disk. pre_loaded_hdf5_file is the loaded hdf5 file as dictionary. If None, the hdf5 file will be read from the disk and consume less RAM.
indices – indices of the subset of repertoires in the data that will be used for this dataset. If ‘None’, all repertoires will be used.
label_name – the name of the label to be predicted
eval_only – whether the dataloader will only be used for evaluation (no training). if false, sample_n_sequences can be set
is_train – whether this is a dataloader for training data. If true, self.training_batch_size is used.
n_workers – the number of workers used in torch.utils.data.DataLoader
- Returns:
a Pytorch dataloader
- store(path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.GradientBoosting module¶
- class immuneML.ml_methods.classifiers.GradientBoosting.GradientBoosting(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodGradient Boosting classifier which wraps scikit-learn’s GradientBoostingClassifier. Input arguments for the method are the same as supported by scikit-learn (see GradientBoostingClassifier scikit-learn documentation for details).
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_gradient_boosting: GradientBoosting: # arguments as defined by scikit-learn n_estimators: 100 learning_rate: 0.1 max_depth: 3 random_state: 42
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
immuneML.ml_methods.classifiers.KNN module¶
- class immuneML.ml_methods.classifiers.KNN.KNN(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodThis is a wrapper of scikit-learn’s KNeighborsClassifier class. This ML method creates a distance matrix using the given encoded data. If the encoded data is already a distance matrix (for example, when using the Distance or CompAIRRDistance encoders), please use PrecomputedKNN instead.
Please see the KNeighborsClassifier scikit-learn documentation of KNeighborsClassifier for the parameters.
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_knn_method: KNN: # sklearn parameters (same names as in original sklearn class) weights: uniform # always use this setting for weights n_neighbors: [5, 10, 15] # find the optimal number of neighbors # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under KNN is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_knn: KNN
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
immuneML.ml_methods.classifiers.KerasSequenceCNN module¶
- class immuneML.ml_methods.classifiers.KerasSequenceCNN.KerasSequenceCNN(units_per_layer: list = None, activation: str = None, training_percentage: float = None)[source]¶
Bases:
MLMethodA CNN-based classifier for sequence datasets. Should be used in combination with
source.encodings.onehot.OneHotEncoder.OneHotEncoder. This classifier integrates the CNN proposed by Mason et al., the original code can be found at: https://github.com/dahjan/DMS_opt/blob/master/scripts/CNN.pyNote: make sure keras and tensorflow dependencies are installed (see installation instructions).
Reference: Derek M. Mason, Simon Friedensohn, Cédric R. Weber, Christian Jordi, Bastian Wagner, Simon M. Men1, Roy A. Ehling, Lucia Bonati, Jan Dahinden, Pablo Gainza, Bruno E. Correia and Sai T. Reddy ‘Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning’. Nat Biomed Eng 5, 600–612 (2021). https://doi.org/10.1038/s41551-021-00699-9
Specification arguments:
units_per_layer (list): A nested list specifying the layers of the CNN. The first element in each nested list defines the layer type, other elements define the layer parameters. Valid layer types are: CONV (keras.layers.Conv1D), DROP (keras.layers.Dropout), POOL (keras.layers.MaxPool1D), FLAT (keras.layers.Flatten), DENSE (keras.layers.Dense). The parameters per layer type are as follows:
[CONV, <filters>, <kernel_size>, <strides>]
[DROP, <rate>]
[POOL, <pool_size>, <strides>]
[FLAT]
[DENSE, <units>]
activation (str): The Activation function to use in the convolutional or dense layers. Activation functions can be chosen from keras.activations. For example, rely or softmax. By default, relu is used.
training_percentage (float): The fraction of sequences that will be randomly assigned to form the training set (the rest will be the validation set). Should be a value between 0 and 1. By default, training_percentage is 0.7.
YAML specification:
definitions: ml_methods: my_cnn: KerasSequenceCNN: training_percentage: 0.7 units_per_layer: [[CONV, 400, 3, 1], [DROP, 0.5], [POOL, 2, 1], [FLAT], [DENSE, 50]] activation: relu
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- check_encoder_compatibility(encoder)[source]¶
Checks whether the given encoder is compatible with this ML method, and throws an error if it is not.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_params(for_refitting=False)[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- load(path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.LogRegressionCustomPenalty module¶
- class immuneML.ml_methods.classifiers.LogRegressionCustomPenalty.LogRegressionCustomPenalty(non_penalized_features: list = None, name: str = None, label: Label = None, non_penalized_encodings: list = None, **kwargs)[source]¶
Bases:
MLMethodLogistic Regression with custom penalty factors for specific features.
Specification arguments:
non_penalized_features (list): List of feature names that should not be penalized.
non_penalized_encodings (list): List of encoding names (class names) whose features should not be penalized. This parameter can be used only in combination with CompositeEncoder. None fo the features from the specified encodings will be penalized. If both non_penalized_features and non_penalized_encodings are provided, the union of the two will be used.
Other supported arguments are inherited from LogitNet of python-glmnet package and will be directly passed to it. n_jobs will be overwritten to use the number of CPUs specified for the instruction (e.g. in TrainMLModel).
YAML specification:
ml_methods: custom_log_reg: LogRegressionCustomPenalty: alpha: 1 n_lambda: 100 non_penalized_features: [] non_penalized_encodings: ['Metadata'] random_state: 42
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_params(for_refitting=False) dict[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- load(path: Path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.LogisticRegression module¶
- class immuneML.ml_methods.classifiers.LogisticRegression.LogisticRegression(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodThis is a wrapper of scikit-learn’s LogisticRegression class. Please see the LogisticRegression scikit-learn documentation of LogisticRegression for the parameters.
Note: if you are interested in plotting the coefficients of the logistic regression model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_logistic_regression: # user-defined method name LogisticRegression: # name of the ML method # sklearn parameters (same names as in original sklearn class) penalty: l1 # always use penalty l1 C: [0.01, 0.1, 1, 10, 100] # find the optimal value for C # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under LogisticRegression is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_logistic_regression: LogisticRegression
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- default_parameters = {'max_iter': 1000, 'solver': 'saga'}¶
immuneML.ml_methods.classifiers.MLMethod module¶
- class immuneML.ml_methods.classifiers.MLMethod.MLMethod(name: str = None, label: Label = None)[source]¶
Bases:
objectML method classifiers are algorithms which can be trained to predict some label on immune repertoires, receptors or sequences.
These methods can be trained using the TrainMLModel instruction, and previously trained models can be applied to new data using the MLApplication instruction.
When choosing which ML method(s) are most suitable for your use-case, please consider the following table:
- DOCS_TITLE = 'Classifiers'¶
- abstract can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- abstract can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- check_encoder_compatibility(encoder)[source]¶
Checks whether the given encoder is compatible with this ML method, and throws an error if it is not.
This method should not be overwritten.
- fit(encoded_data: EncodedData, label: Label, optimization_metric: str = None, cores_for_training: int = 2)[source]¶
The fit method is called by the MLMethodTrainer to fit a model to a specific encoded dataset. This method internally calls methods to initialise and perform model fitting.
This method should not be overwritten.
- fit_by_cross_validation(encoded_data: EncodedData, label: Label, optimization_metric, number_of_splits: int = 5, cores_for_training: int = 2)[source]¶
See also: _fit_by_cross_validation.
This method should not be overwritten.
- abstract get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_package_info() str[source]¶
Should return the relevant version numbers of immuneML and all external packages that were used for the immuneML implementation. This information will be exported and is required for transparency and reproducibility.
This method should be overwritten to add any additional packages if necessary. For instance, versions of scikit-learn.
- abstract get_params(for_refitting=False) dict[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- abstract load(path: Path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- predict(encoded_data: EncodedData, label: Label)[source]¶
Safely calls ‘_predict’ after checking the label is matching. This method should not be overwritten.
- predict_proba(encoded_data: EncodedData, label: Label)[source]¶
Safely calls ‘_predict_proba’ after checking the label is matching. This method should not be overwritten.
- abstract store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.PrecomputedKNN module¶
- class immuneML.ml_methods.classifiers.PrecomputedKNN.PrecomputedKNN(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodThis is a wrapper of scikit-learn’s KNeighborsClassifier class. This ML method takes a pre-computed distance matrix, as created by the Distance or CompAIRRDistance encoders. If you would like to use a different encoding in combination with KNN, please use KNN instead.
Please see the KNN scikit-learn documentation of KNeighborsClassifier for the parameters.
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_knn_method: PrecomputedKNN: # sklearn parameters (same names as in original sklearn class) weights: uniform # always use this setting for weights n_neighbors: [5, 10, 15] # find the optimal number of neighbors # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under KNN is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_knn: PrecomputedKNN
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
immuneML.ml_methods.classifiers.ProbabilisticBinaryClassifier module¶
- class immuneML.ml_methods.classifiers.ProbabilisticBinaryClassifier.ProbabilisticBinaryClassifier(max_iterations: int, update_rate: float, likelihood_threshold: float)[source]¶
Bases:
MLMethodProbabilisticBinaryClassifier predicts the class assignment in binary classification case based on encoding examples by number of successful trials and total number of trials. It models this ratio by one beta distribution per class and predicts the class of the new examples using log-posterior odds ratio with threshold at 0.
ProbabilisticBinaryClassifier is based on the paper (details on the classification can be found in the Online Methods section): Emerson, Ryan O., William S. DeWitt, Marissa Vignali, Jenna Gravley, Joyce K. Hu, Edward J. Osborne, Cindy Desmarais, et al. ‘Immunosequencing Identifies Signatures of Cytomegalovirus Exposure History and HLA-Mediated Effects on the T Cell Repertoire’. Nature Genetics 49, no. 5 (May 2017): 659–65. doi.org/10.1038/ng.3822.
Specification arguments:
max_iterations (int): maximum number of iterations while optimizing the parameters of the beta distribution (same for both classes)
update_rate (float): how much the computed gradient should influence the updated value of the parameters of the beta distribution
likelihood_threshold (float): at which threshold to stop the optimization (default -1e-10)
YAML specification:
definitions: ml_methods: my_probabilistic_classifier: # user-defined name of the ML method ProbabilisticBinaryClassifier: # method name max_iterations: 1000 update_rate: 0.01
- SMALL_POSITIVE_NUMBER = 1e-15¶
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_params(for_refitting=False)[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- load(path: Path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.RandomForestClassifier module¶
- class immuneML.ml_methods.classifiers.RandomForestClassifier.RandomForestClassifier(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodThis is a wrapper of scikit-learn’s RandomForestClassifier class. Please see the RandomForestClassifier scikit-learn documentation of RandomForestClassifier for the parameters.
Note: if you are interested in plotting the coefficients of the random forest classifier model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_random_forest_classifier: # user-defined method name RandomForestClassifier: # name of the ML method # sklearn parameters (same names as in original sklearn class) random_state: 100 # always use this value for random state n_estimators: [10, 50, 100] # find the optimal number of trees in the forest # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under RandomForestClassifier is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_random_forest: RandomForestClassifier
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
immuneML.ml_methods.classifiers.ReceptorCNN module¶
- class immuneML.ml_methods.classifiers.ReceptorCNN.ReceptorCNN(kernel_count: int = None, kernel_size=None, positional_channels: int = None, sequence_type: str = None, device=None, number_of_threads: int = None, random_seed: int = None, learning_rate: float = None, iteration_count: int = None, l1_weight_decay: float = None, l2_weight_decay: float = None, batch_size: int = None, training_percentage: float = None, evaluate_at: int = None, background_probabilities=None)[source]¶
Bases:
MLMethodA CNN which separately detects motifs using CNN kernels in each chain of paired receptor data, combines the kernel activations into a unique representation of the receptor and uses this representation to predict the antigen binding.
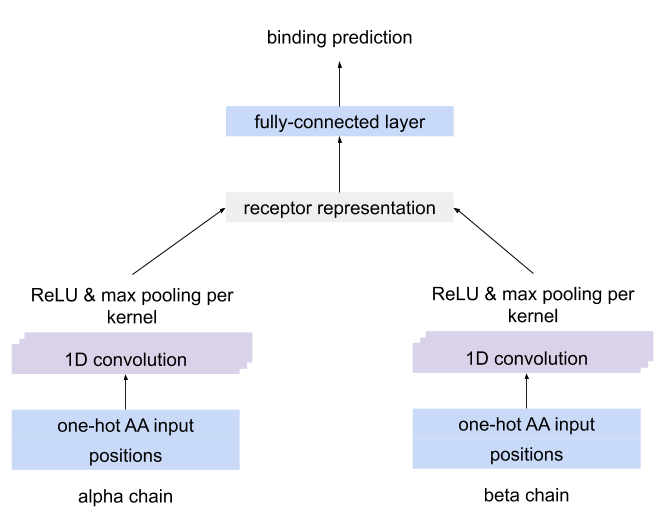
The architecture of the CNN for paired-chain receptor data¶
Requires one-hot encoded data as input (as produced by OneHot encoder), where use_positional_info must be set to True.
Notes:
ReceptorCNN can only be used with ReceptorDatasets, it does not work with SequenceDatasets
ReceptorCNN can only be used for binary classification, not multi-class classification.
Specification arguments:
kernel_count (count): number of kernels that will look for motifs for one chain
kernel_size (list): sizes of the kernels = how many amino acids to consider at the same time in the chain sequence, can be a tuple of values; e.g. for value [3, 4] of kernel_size, kernel_count*len(kernel_size) kernels will be created, with kernel_count kernels of size 3 and kernel_count kernels of size 4 per chain
positional_channels (int): how many positional channels where included in one-hot encoding of the receptor sequences (OneHot encoder adds 3 positional channels positional information is enabled)
sequence_type (SequenceType): type of the sequence
device: which device to use for the model (cpu or gpu) - for more details see PyTorch documentation on device parameter
number_of_threads (int): how many threads to use
random_seed (int): number used as a seed for random initialization
learning_rate (float): learning rate scaling the step size for optimization algorithm
iteration_count (int): for how many iterations to train the model
l1_weight_decay (float): weight decay l1 value for the CNN; encourages sparser representations
l2_weight_decay (float): weight decay l2 value for the CNN; shrinks weight coefficients towards zero
batch_size (int): how many receptors to process at once
training_percentage (float): what percentage of data to use for training (the rest will be used for validation); values between 0 and 1
evaluate_at (int): when to evaluate the model, e.g. every 100 iterations
background_probabilities: used for rescaling the kernel values to produce information gain matrix; represents the background probability of each amino acid (without positional information); if not specified, uniform background is assumed
YAML specification:
definitions: ml_methods: my_receptor_cnn: ReceptorCNN: kernel_count: 5 kernel_size: [3] positional_channels: 3 sequence_type: amino_acid device: cpu number_of_threads: 16 random_seed: 100 learning_rate: 0.01 iteration_count: 10000 l1_weight_decay: 0 l2_weight_decay: 0 batch_size: 5000
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- check_encoder_compatibility(encoder)[source]¶
Checks whether the given encoder is compatible with this ML method, and throws an error if it is not.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_params(for_refitting=False)[source]¶
Returns the model parameters and their values in a readable yaml-friendly way (a dictionary consisting of ints, floats, strings, lists and dictionaries). This may simply be vars(self), but if an internal (sklearn) model is fitted, the parameters of the internal model should be included as well.
- Parameters:
for_refitting
- load(path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.SVC module¶
- class immuneML.ml_methods.classifiers.SVC.SVC(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodThis is a wrapper of scikit-learn’s LinearSVC class. Please see the LinearSVC scikit-learn documentation of SVC for the parameters.
Note: if you are interested in plotting the coefficients of the SVC model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_svc: # user-defined method name SVC: # name of the ML method # sklearn parameters (same names as in original sklearn class) C: [0.01, 0.1, 1, 10, 100] # find the optimal value for C # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under SVC is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_svc: SVC
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
immuneML.ml_methods.classifiers.SVM module¶
- class immuneML.ml_methods.classifiers.SVM.SVM(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
SklearnMethodThis is a wrapper of scikit-learn’s SVC class. Please see the SVC scikit-learn documentation of SVC for the parameters.
Note: if you are interested in plotting the coefficients of the SVM model, consider running the Coefficients report.
For usage instructions, check
SklearnMethod.YAML specification:
definitions: ml_methods: my_svm: # user-defined method name SVM: # name of the ML method # sklearn parameters (same names as in original sklearn class) C: [0.01, 0.1, 1, 10, 100] # find the optimal value for C kernel: linear # Additional parameter that determines whether to print convergence warnings show_warnings: True # if any of the parameters under SVM is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5 # alternative way to define ML method with default values: my_default_svm: SVM
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
immuneML.ml_methods.classifiers.SklearnMethod module¶
- class immuneML.ml_methods.classifiers.SklearnMethod.SklearnMethod(parameter_grid: dict = None, parameters: dict = None)[source]¶
Bases:
MLMethodBase class for ML methods imported from scikit-learn. The classes inheriting SklearnMethod acting as wrappers around imported ML methods from scikit-learn have to implement: - the __init__() method, - get_params(label) and - _get_ml_model()
Other methods can also be overwritten if needed. The arguments and specification described bellow applied for all classes inheriting SklearnMethod.
Specification arguments:
parameters: a dictionary of parameters that will be directly passed to scikit-learn’s class upon calling __init__() method; for detailed list see scikit-learn’s documentation of the specific class inheriting SklearnMethod
parameter_grid: a dictionary of parameters which all have to be valid arguments for scikit-learn’s corresponding class’ __init__() method (same as parameters), but unlike parameters argument can contain list of values instead of one value; if this is specified and “model_selection_cv” is True (in the specification) or just if fit_by_cross_validation() is called, a grid search will be performed over these parameters and the optimal model will be kept
YAML specification:
- definitions:
- ml_methods:
- ml_methods:
- log_reg:
- LogisticRegression: # name of the class inheriting SklearnMethod
# sklearn parameters (same names as in original sklearn class) max_iter: 1000 # specific parameter value penalty: l1 # Additional parameter that determines whether to print convergence warnings show_warnings: True
# if any of the parameters under LogisticRegression is a list and model_selection_cv is True, # a grid search will be done over the given parameters, using the number of folds specified in model_selection_n_folds, # and the optimal model will be selected model_selection_cv: True model_selection_n_folds: 5
- svm_with_cv:
- SVM: # name of another class inheriting SklearnMethod
# sklearn parameters (same names as in original sklearn class) alpha: 10 # Additional parameter that determines whether to print convergence warnings show_warnings: True
# no grid search will be done model_selection_cv: False
- FIT = 'fit'¶
- FIT_CV = 'fit_CV'¶
- apply_with_weights(method, weights, **kwargs)[source]¶
Can be used to run self.model.fit, self.model.predict or self.model.predict_proba with sample weights if supported
- Parameters:
method – self.model.fit, self.model.predict or self.model.predict_proba
- Returns:
the result of the supplied method
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.
- get_compatible_encoders()[source]¶
Returns a list of compatible encoders. This method should be overwritten for every MLMethod.
for example:
from immuneML.encodings.evenness_profile.EvennessProfileEncoder import EvennessProfileEncoder
return [EvennessProfileEncoder]
- get_package_info() str[source]¶
Should return the relevant version numbers of immuneML and all external packages that were used for the immuneML implementation. This information will be exported and is required for transparency and reproducibility.
This method should be overwritten to add any additional packages if necessary. For instance, versions of scikit-learn.
- abstract get_params(for_refitting=False)[source]¶
Returns the model parameters in a readable yaml-friendly way (consisting of lists, dictionaries and strings).
- Parameters:
for_refitting
- load(path: Path)[source]¶
The load function can load the model given the folder where the same class of the model was previously stored using the store function. It reads in the parameters of the model and sets the values to the object attributes so that the model can be reused. For instance, this is used in MLApplication instruction when the previously trained model is applied on a new dataset.
- Parameters:
path (Path) – path to the folder where the model was stored using store() function
- Returns:
it does not have a return value, but sets the attribute values of the object instead
- store(path: Path)[source]¶
The store function stores the object on which it is called so that it can be imported later using load function. It typically uses pickle, yaml or similar modules to store the information. It can store one or multiple files.
- Parameters:
path (Path) – path to folder where to store the model
- Returns:
it does not have a return value
immuneML.ml_methods.classifiers.TCRdistClassifier module¶
- class immuneML.ml_methods.classifiers.TCRdistClassifier.TCRdistClassifier(percentage: float, show_warnings: bool = True)[source]¶
Bases:
SklearnMethodImplementation of a nearest neighbors classifier based on TCR distances as presented in Dash P, Fiore-Gartland AJ, Hertz T, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017; 547(7661):89-93. doi:10.1038/nature22383.
This method is implemented using scikit-learn’s KNeighborsClassifier with k determined at runtime from the training dataset size and weights linearly scaled to decrease with the distance of examples.
Specification arguments:
percentage (float): percentage of nearest neighbors to consider when determining receptor specificity based on known receptors (between 0 and 1)
show_warnings (bool): whether to show warnings generated by scikit-learn, by default this is True.
YAML specification:
definitions: ml_methods: my_tcr_method: TCRdistClassifier: percentage: 0.1 show_warnings: True
- can_fit_with_example_weights() bool[source]¶
Returns a boolean value indicating whether the model can be fit with example weights. Example weights allow to up-weight the importance of certain examples, and down-weight the importance of others.
- can_predict_proba() bool[source]¶
Returns whether the ML model can be used to predict class probabilities or class assignment only. This method should be overwritten to return True if probabilities can be predicted, and False if they cannot.